Extended Data Figure 1. CRISPR-Cas9-based screen identifying Ldlrad3 as required factor for VEEV infectivity.
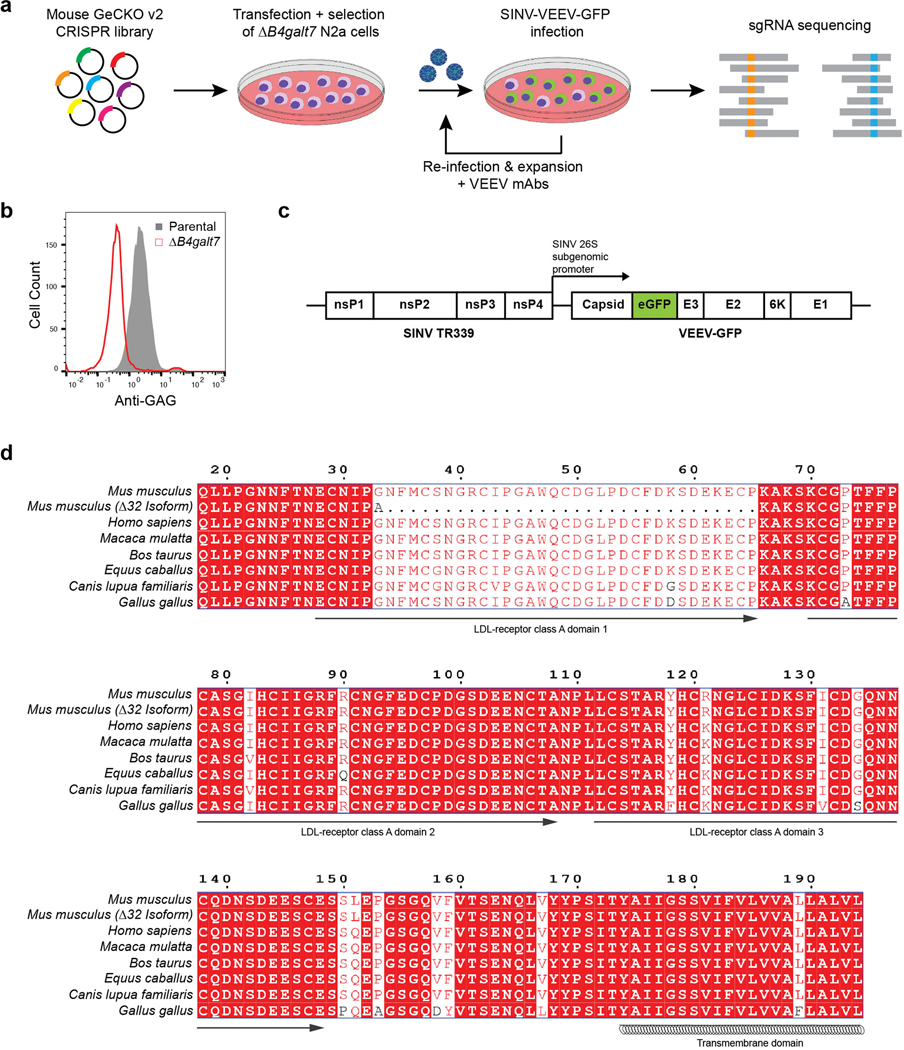
a. ΔB4galt7 N2a cells were transfected separately with two half libraries containing 130,209 sgRNAs, puromycin selected, and then inoculated with SINV-VEEV-GFP (TrD strain) at an MOI of 1. After 24 h, GFP-negative cells were sorted, expanded in the presence of anti-VEEV mAbs (VEEV-57, VEEV-67, and VEEV-68 [2 μg/ml]), and re-inoculated with SINV-VEEV-GFP. The infection and sorting process were repeated twice. Genomic DNA from GFP-negative cells was sequenced for sgRNA abundance. b. Representative flow cytometry histogram of parental N2a (gray) and ΔB4galt7 N2a (red) cells stained for heparan sulfate (HS) surface expression using R1725, a rodent herpesvirus immune evasion protein that binds to HS. c. Schematic diagram of chimeric SINV-VEEV virus. The chimera contains the non-structural genes from SINV (strain TR339), structural genes from VEEV (IAB strain TrD, IC strain INH9813, or ID strain ZPC738), and an eGFP gene (green) between the capsid and E3 protein. The insertion of GFP has minimal effects on virus infection and replication20,24. d. Sequence alignment of mouse (Mus musculus), mouse Δ32 N-terminus isoform, human (Homo sapiens), rhesus macaque (Macaca mulatta), cattle (Bos taurus), horse (Equus caballus), dog (Canis lupus familiaris), and chicken (Gallus gallus) Ldlrad3 ectodomain using ESPript 3. Red boxes indicate conserved residues between orthologs. The predicted domains based on sequence similarity to other related proteins and the transmembrane domain are indicated below the sequence.
