Figure 5. Engineering of chimeric hES9S-humanized ribosomes in yeast.
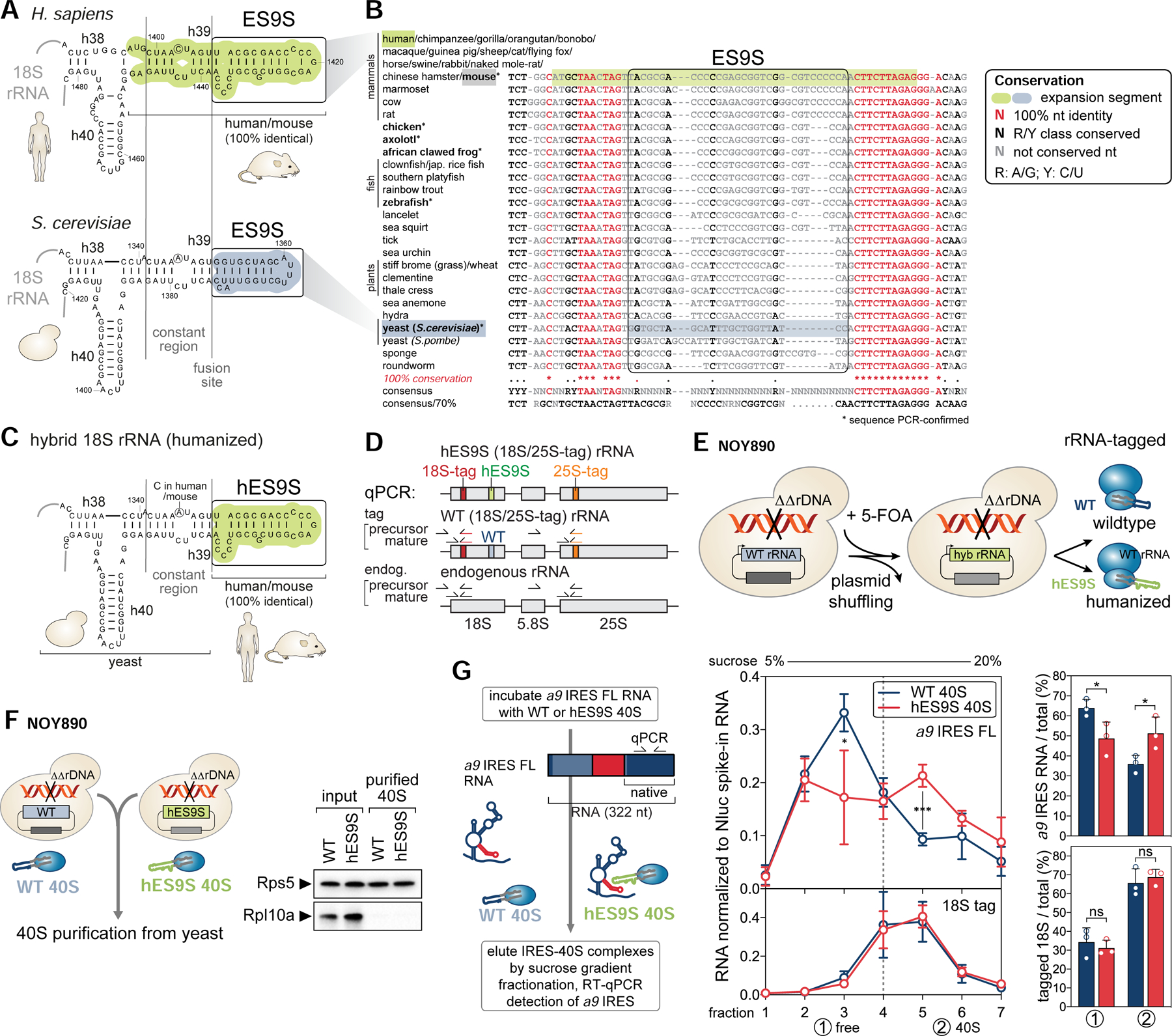
(A) Secondary structure models of the human (green) and yeast (blue) 18S rRNA region containing ES9S. The constant region (h39) and ES9S-fusion site selected for chimeric 18S rRNA engineering are indicated.
(B) A 40-way multiple sequence alignment (MSA) and conservation analysis of the highly conserved 18S rRNA region in which the more variable ES9S is embedded. Nts are color coded according to conservation. For six species (bold, asterisk) annotated 18S rRNA sequences were confirmed by RT-PCR spanning the ES9S region. R, purine; Y, pyrimidine.
(C) Structure model of the engineered yeast 18S rRNA after exchange of the yeast to human ES9S (hES9S, green).
(D) The rRNA cassette that encodes 18S, 5.8S and 25S rRNA indicating the position of unique sequence tags in 18S (red) and 25S (orange) rRNA used for RT-qPCR to detect precursor and mature forms of endogenous and tagged yeast rRNA.
(E) The plasmid shuffling approach to generate yeast strains that contain a homozygous knock-out of the rDNA locus (NOY890) and exclusively express plasmid-encoded tagged chimeric 18S rRNA as in (C). See also Figure S5.
(F) 40S subunits of WT and hES9S yeast strains (NOY890) were purified by sequential sucrose gradient sedimentation (see also Figure S6). The purity of the isolated 40S was confirmed by WB analysis of RPs compared to the input lysate. RPL10A/uL1 is yeast Rpl1 (referred to as Rpl10a).
(G) In vitro binding assays using purified WT and hES9S 40S subunits to test direct binding to the a9 IRES FL RNA. IRES-40S complexes were eluted by 5–20% sucrose gradient fractionation. Co-sedimentation of 40S (18S rRNA tag) and bound RNA (a9 IRES FL) was detected by RT-qPCR, normalized to a Nluc spike-in RNA (average ± SD, n = 3). IRES-40S co-sedimentation was assessed by integrating the gradient distribution and expressed for the free (1) and 40S (2) fractions as the percentage relative to the total (average ± SD, n = 3).
