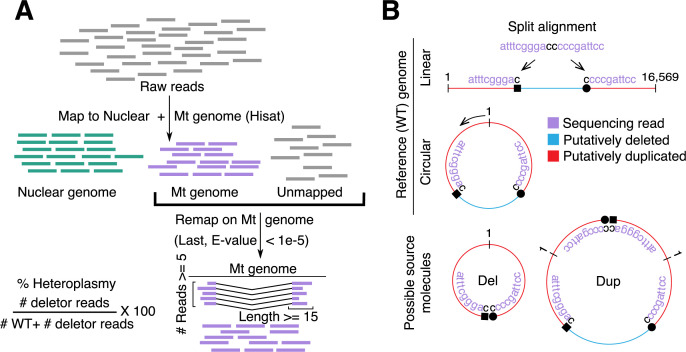
Fig 1. MitoSAlt pipeline overview.
(A) Raw sequencing reads are mapped first to the nuclear and mitochondrial (Mt) genomes using a fast aligner, followed by precision alignment of unmapped and Mt mapped reads to the Mt genome to identify “split” reads informative of structural breakpoints. (B) Dual interpretation of split alignments: a split read can represent either a deletion or a complementary arc duplication, and these scenarios are indistinguishable using short-read sequencing.