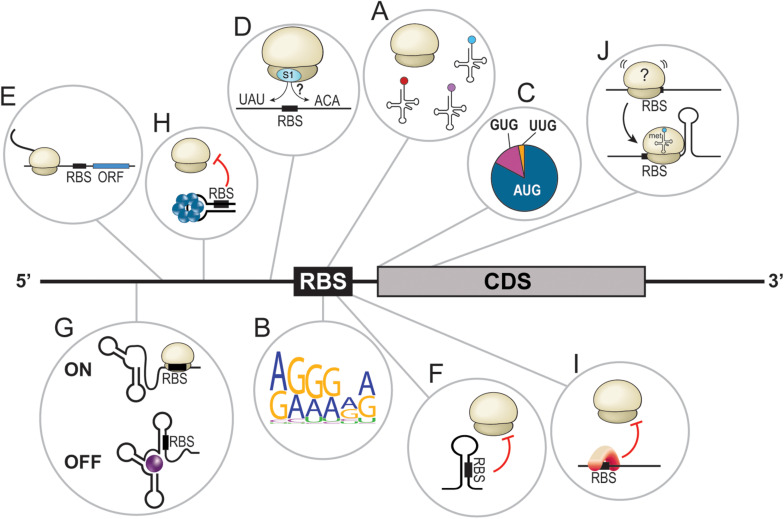
FIGURE 1.
Overview of bacterial translational determinants. (A) Proper assembly of the translational machinery, including the ribosomes and tRNAs, is required for efficient translation. (B) Graphical representation (Crooks et al., 2004) of the Shine-Dalgarno core sequence in E. coli based on 50 random mRNAs. (C) Distribution of the different initiator codons in bacteria. AUG = 80%, GUG = 12%, and UUG = 8%. (D) Translation enhancers are often A/U- or C/A-rich sequences located upstream or downstream of the ribosome binding site (RBS). (E) Leader open reading frames dictate the translation of downstream CDS. (F) Secondary structures in the 5′UTR sequester the RBS and hinder translation by steric inhibition. (G) Riboswitches are complex RNA structures in 5′UTRs that, upon binding of a ligand, change conformation to turn the translation of the downstream open reading frame ON or OFF. (H) The binding of a protein forces the adoption of a 5′UTR structure that sequesters the RBS, preventing translation initiation. (I) Binding of a protein to the RBS prevents translation initiation. (J) A stem loop at the beginning of the coding sequence seems to stabilize the ribosome at the RBS and facilitate translation initiation.