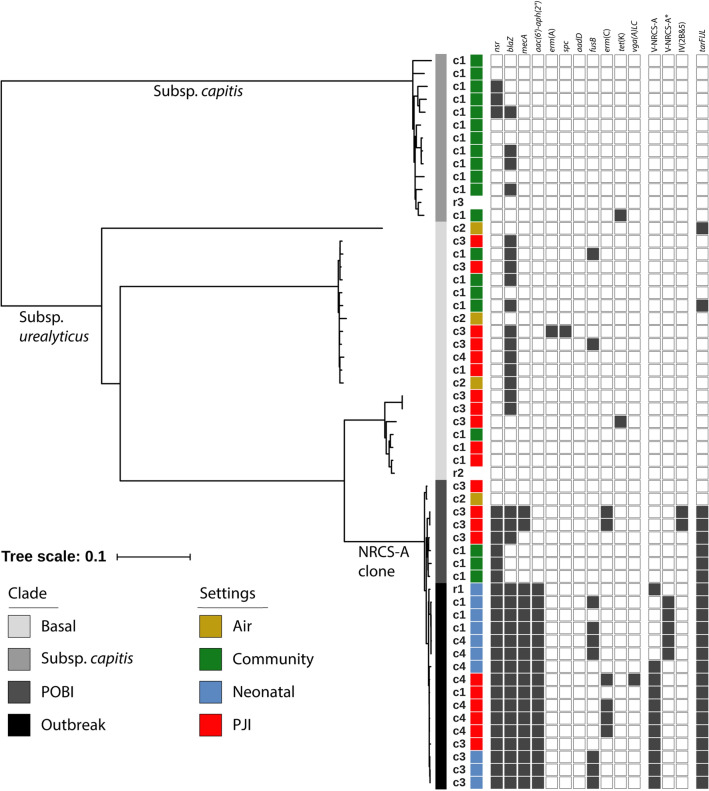
Figure 2.
Midpoint-rooted maximum-likelihood phylogeny of 56 S. capitis isolates and presence of resistance genes based on a 75% core genome. Different centres are identified as c1–c4 and reference isolates named r1 (NRCS-A prototype strain CR01), r2 (CCUG 55892), and r3 (CCUG 35173). All isolates except r1 were isolated in Sweden. The settings where all Swedish strains were isolated are presented in colour: yellow = air, green = community, blue = neonatal, red = PJI, and white = no data. From c1 was included nasal isolates (n = 20), PJI isolates (n = 4) and NICU isolates (n = 3), from c2 operating theatre air (n = 4), from c3 PJI isolates (n = 12) and NICU isolates (n = 3) and from c4 PJI isolates (n = 5) and NICU isolates (n = 3). The phylogeny highlighted the subsp. capitis, Basal, and NRCS-A clade sublineages. Black blocks represent presence of genes mediating antibiotic resistance and SCCmec type.