Figure 6: Genetic inactivation of Klf4 specifically in SMC-P does not result in marked changes in the distribution of cells within perivascular clusters but impacted transcriptomic clusters of MФs and endothelial cells.
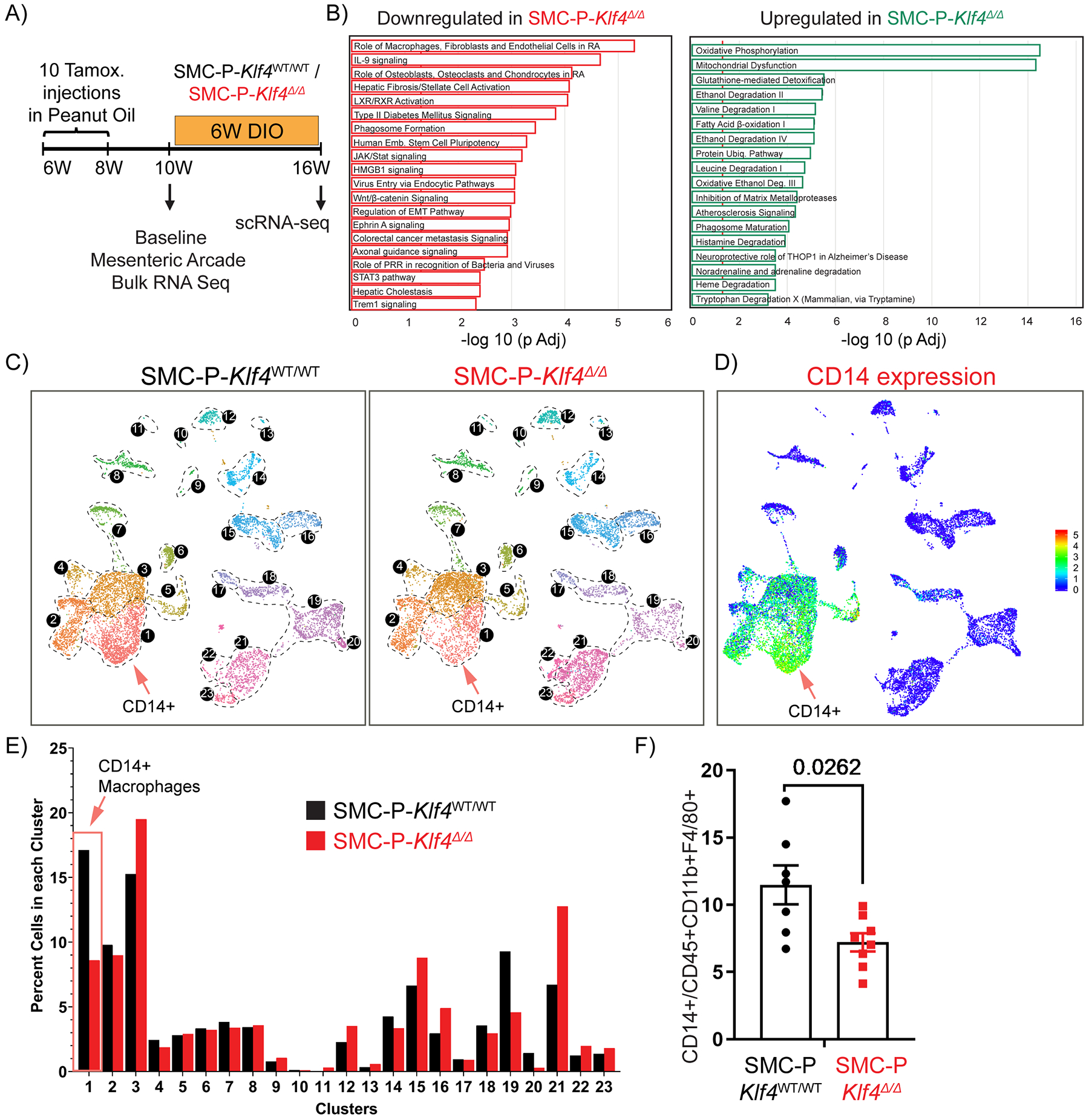
(A) Schematic of experimental design. SMC-P Klf4WT/WT or SMC-P Klf4Δ/Δ mice were injected with tamoxifen in peanut oil and following a 2-week washout period either tissues were harvested for baseline bulk RNA-sequencing experiment or mice fed DIO diet for 6 weeks before subsequent scRNAseq. (B) Mesenteric arcades, including arteries of variable sizes and surrounding adipose tissue (excluding intestines), were harvested, and SVF cells were prepared for total RNA isolation. Bulk RNA-seq and Ingenuity Pathway Analysis were performed. Significantly enriched pathways were identified using a 5% false discovery rate cutoff. Data are presented as pathways downregulated or upregulated in SMC-P Klf4Δ/Δ mice when compared to SMC-P Klf4WT/WT mice. Enrichment is shown as −log10 of Padj values. (C) UMAP plots showing integrated Seurat libraries depicting 23 clusters. Four libraries were included in the analysis: eYFP+CD45‒, eYFP+CD45+CD11b+F4/80+ (eYFP+ MФ), eYFP‒CD45+CD11b+F4/80+ (eYFP‒ MФ), and eYFP‒CD45‒ from 4 different conditions as in Figure 2. Cells from the libraries from 6-weeks DIO diet-fed SMC-P Klf4WT/WT mice (left panel) and SMC-P Klf4Δ/Δ mice (right panel) are color-coded by clusters as described in Figure 2. (D) Feature plot for Cd14, one of the top significantly differentially expressed genes in cluster 1. (E) Quantitative analysis for frequency distribution of cells from SMC-P Klf4WT/WT and SMC-P Klf4Δ/Δ mice among different clusters not-segregated based on the library of flow sorting origin. (F) SMC-P Klf4WT/WT (n = 7) and SMC-P Klf4Δ/Δ mice (n = 8) were fed tamoxifen in diet and following a 2-week washout were fed a high fat diet for six weeks. Epididymal adipose tissue SVF cell suspensions were stained with antibodies for CD14. The frequency of CD14+ MФs among CD45+CD11b+F4/80+ MФs was plotted as mean ± SEM. An unpaired t-test with Welch’s correction yielded P = 0.0262.
