Figure 2. Allele default assignment strategy used by many testing platforms.
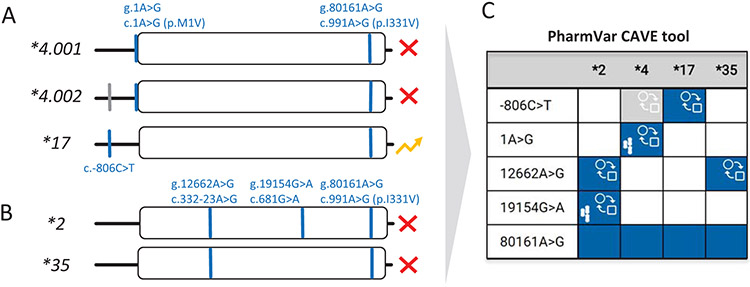
Many pharmacogenetic test platforms only comprise the more commonly observed SNVs and do not include all known SNVs, or star (*) alleles that are defined by PharmVar. Consequently, some alleles may not be identified or receive an assignment by ‘default’. It is therefore important to know which SNVs are tested in order to fully understand how alleles were called and translated into phenotype, as well as to appreciate a test’s limitations. Panel A visualizes the two known CYP2C19*4 subvariants which are nonfunctional due to c.1A>G (p.M1V). The CYP2C19*4.002 subvariant also carries c.−806C>T which is the core SNV defining the increased function *17 allele. Thus, if testing only includes c.−806C>T and not c.1A>G, a CYP2C19*4 allele will be called *17 potentially leading to an incorrect phenotype assignment. Panel B illustrates that g.12662A>G (c.332-23A>G), which causes aberrant splicing, occurs on both, CYP2C19*2 and *35. If not tested, the latter will be defaulted to a CYP2C19*1 assignment that may also lead to an incorrect phenotype assignment. Panel C shows the comparison of CYP2C19*2, *4, *17 and *35, created with the PharmVar CAVE tool. Blue boxes indicate the presence of a core SNV on all suballeles, while the gray box indicates that the core SNV is not present on all suballeles. The function ( )symbol indicates that a core SNV alters function and the PharmVar (
)symbol indicates that a core SNV alters function and the PharmVar ( ) symbol highlights that a core SNV is unique to a star allele. SNV positions refer to genomic coordinates on the NG_008384.3 reference sequence (with the ATG start codon being +1).
) symbol highlights that a core SNV is unique to a star allele. SNV positions refer to genomic coordinates on the NG_008384.3 reference sequence (with the ATG start codon being +1).