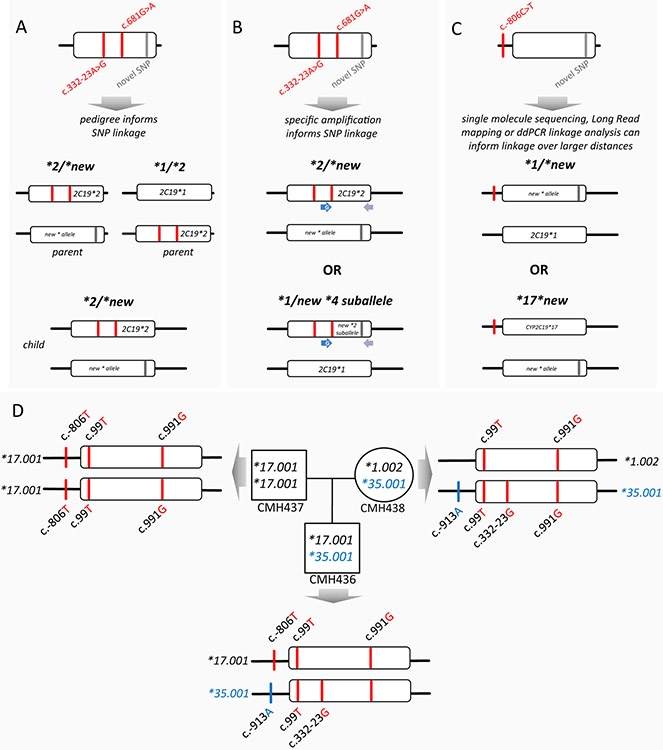
Figure 4. Experimental approaches for phasing SNVs to establish haplotype.
Panels A and B depict a subject who is heterozygous for three SNVs of which two designate the CYP2C19*2 allele. Whether the novel SNV is in cis or trans with the CYP2C19*2 SNVs can be informed by e.g., inheritance (panel A), or experimentally determined using e.g., allele-specific (long-range) PCR, followed by sequencing (panel B), given that the distance among the novel SNV is not exceeding PCR amplification limitations. Panel C provides an example of a subject who is heterozygous for two SNPs, one of which designates the CYP2C19*17 allele. In this instance, the phase of the novel SNV may be inferred by inheritance (not shown) or experimentally determined by single molecule sequencing or utilizing ddPCR-based SNP-linkage analysis. Panel D details a submission for the CYP2C19*35 allele. The haplotype of the CYP2C19*35 suballele was determined from whole genome sequence data of each member of the trio. This data revealed an additional SNV on the CYP2C19*35 haplotype, as well as provided evidence to support changing the allele’s evidence level to ‘Def’.