Figure 5 – P. aeruginosa type I-F1 Cascade can utilize heterologous I-F3 CRISPR arrays in a plasmid interference assay, but mismatches and I-F3b atypical guide RNAs allow privatization.
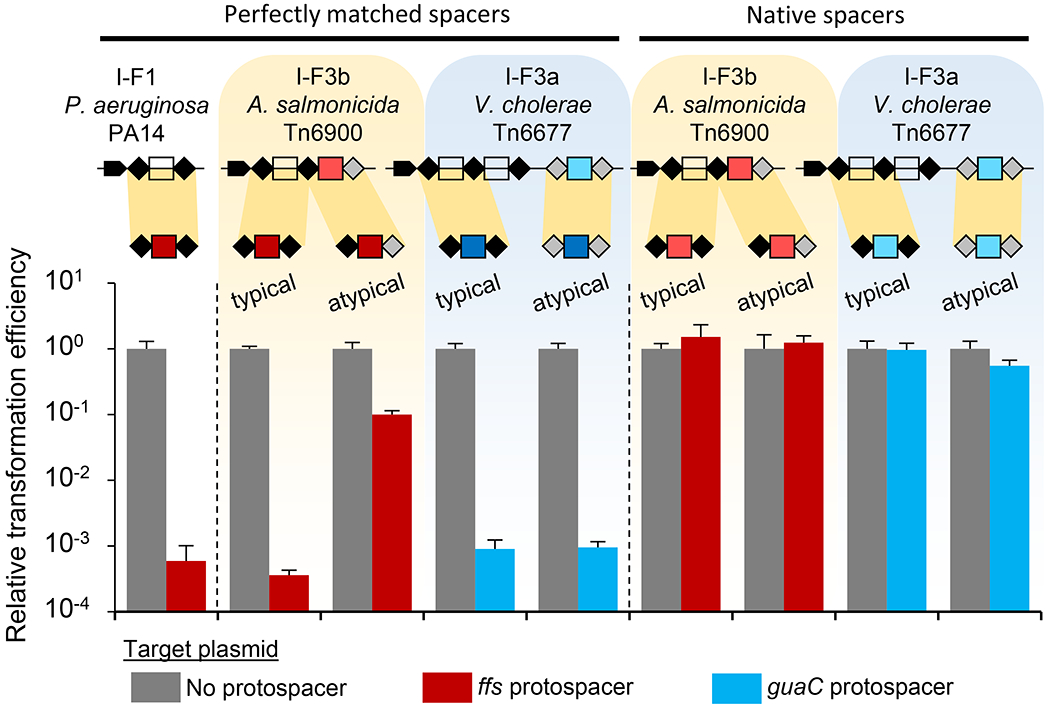
Expression of P. aeruginosa Cas proteins with various arrays reduces transformation efficiency for protospacer containing plasmid, but not control. Single unit arrays from P. aeruginosa PA14 and A. salmonicida S44 Tn6900 with ffs spacer, and V. cholerae HE-45 Tn6677 with guaC spacer were tested in typical and atypical repeat configurations as indicated. Spacers were either perfectly matched to protospacer or contained native mismatches. Repeat configuration sequences are presented in Supplemental Figure S3b. All data indicate mean +/− standard deviation (n=3).
