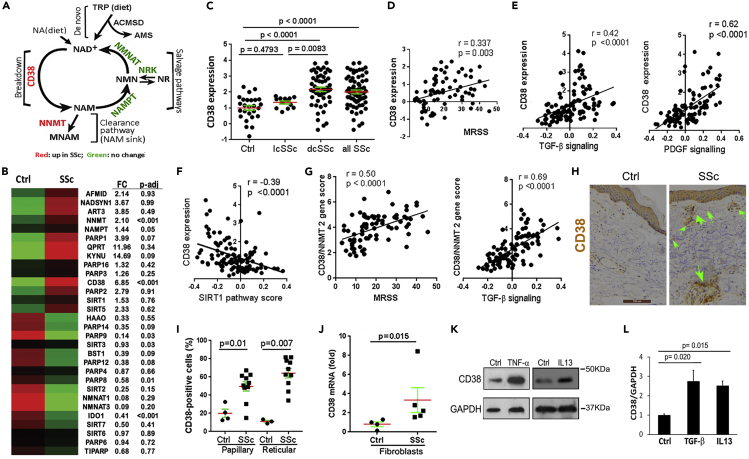
Figure 1.
NAD+-consuming enzyme CD38 significantly elevated in SSc skin biopsies
(A) Schematic of cellular NAD+ metabolism, showing key enzymes catalyzing NAD+ consumption and salvage.
(B) Heatmap indicating differential expression of key enzymes catalyzing NAD+ consumption and salvage in healthy control (n = 26) and SSc (n = 68) skin biopsies (GSE76886, red indicates high and green indicates low expression).
(C) CD38 mRNA expression in healthy control and limited and diffuse cutaneous SSc (lcSSc and dcSSc) skin biopsies.
(D–F) Correlation of CD38 mRNA levels with modified Rodnan skin score (MRSS, range 0–51) and with TGF-β, PDGF, and SIRT1 pathway scores in the skin.
(G) Combined CD38 and NNMT gene score correlation with MRSS and with TGF-β signaling. Pearson's correlation.
(H) Immunohistochemistry of skin biopsies using antibodies to CD38; representative images. Arrows indicate CD38+ cells. Scale bar length represents 100 μm.
(I) Quantitation of CD38+ cells in the dermis; horizontal bars, means ± SEM (standard error of the mean) (10 SSc and 4 healthy control biopsies for (H) and (I), clinical data in Table S1 in Transparent Method).
(J) CD38 mRNA levels in explanted SSc (n = 5) and healthy control (n = 4) fibroblasts determined by qPCR.
(K and L) CD38 in human skin fibroblasts. Confluent cultures were incubated for 24 h with TNF-α (10 ng/mL), IL-13 (10 ng/mL), and TGF-β (10 ng/mL). (K) Whole cell lysates were analyzed by immunoblotting. (L) mRNA levels determined by qPCR, mean ± SEM. Experiments were repeated twice.
AMS: alpha-aminomuconate semialdehyde; ACMSD: aminocarboxymuconate semialdehyde decarboxylase; MNAM: N1-methylnicotinamide; NA: nicotinic acid; NAD: nicotinamide adenine dinucleotide; NAM: nicotinamide; NAMPT: nicotinamide phosphoribosyltransferase; NMN: nicotinamide mononucleotide; NMNAT: nicotinamide mononucleotide adenylyltransferase; NNMT: nicotinamide N-methyltransferase; NR: nicotinamide riboside; NRK: nicotinamide riboside kinase; TRP: tryptophan.