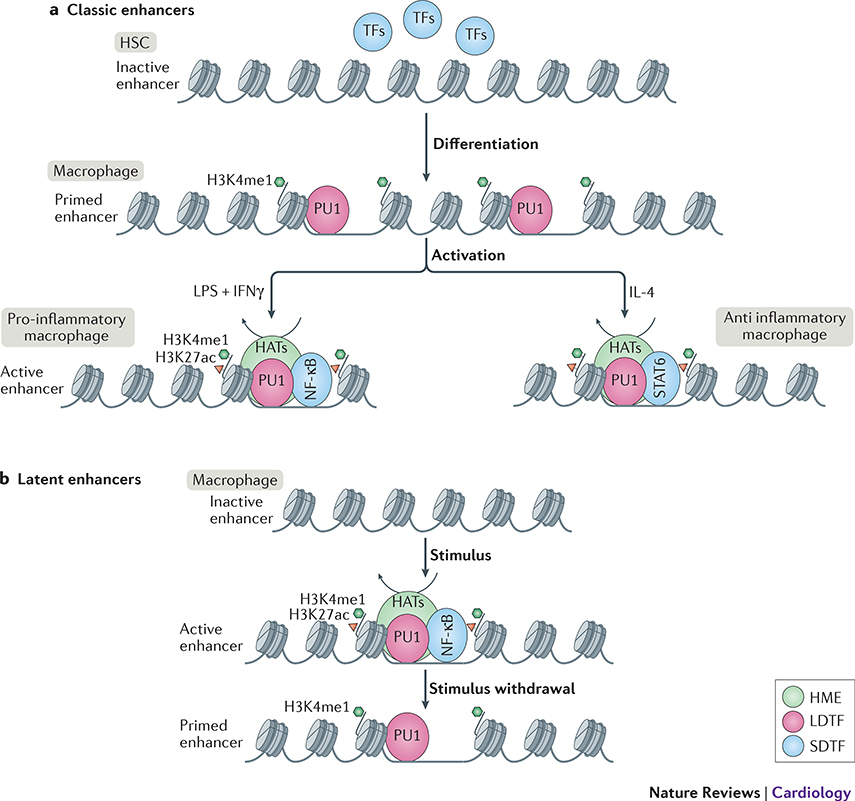
Figure 2: Macrophage enhancer selection. Classic enhancers:
are opened by lineage determining transcription factor (LDTF) PU1 during myeloid lineage differentiation to macrophages. Enhancers are primed by deposition of H3K4me1 by histone modifying enzymes (HMEs) from the lysine methyl transferase (KMT) families 2, 3 or 7 (Box 2). Upon activation, signal dependent transcription factor (SDTFs) such as NF-κB for pro-inflammatory M1-like activation or STAT6 for anti-inflammatory M2-like activation, are recruited to the primed enhancers. This in turn leads to recruitment of histone acetyl transferases (HATs) to set up a permissive chromatin landscape and activate the enhancer. Latent enhancers: are unbound by LDTFs such as PU1 in a steady-state macrophage. Upon stimulus, SDTFs such as NF-κB can bind to their partially uncovered motifs and open up the chromatin, leading to PU1, KMT, and HAT recruitment to fully open up the chromatin and activate the enhancer. After removal of the stimulus, the SDTF leaves the chromatin, and activating histone marks are removed by recruitment of histone de-acetylases (HDACs), however H3K4me1 remains unremoved and keeps enhancer in a primed state for subsequent activation. This enhancer memory enables a quicker activation upon the next encounter of the same stimulus.