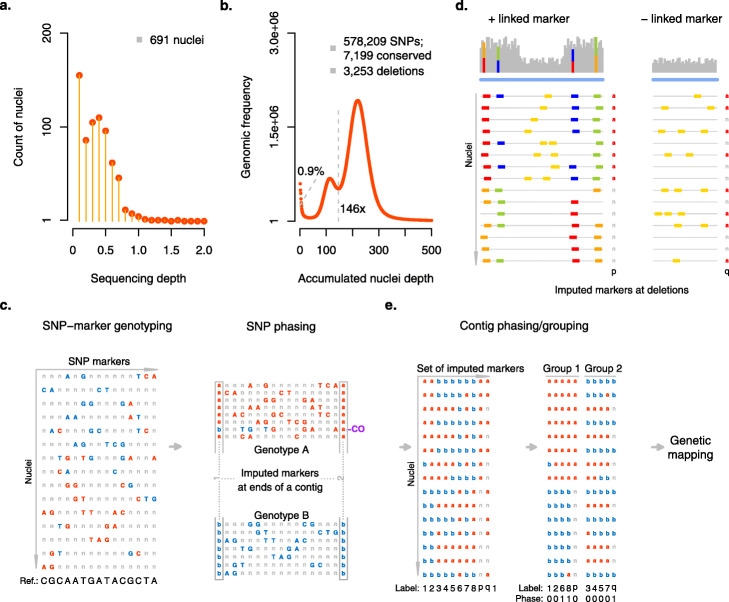
Fig. 2.
Single-pollen nuclei sequencing, variant phasing and genetic mapping. a Sequencing depths of 691 pollen nuclei. b Sequencing depth histogram of pooled pollen short reads. The left-most peak revealed 0.9% of the genome that were not well covered in the pollen read sets (i.e., ≤ 5x). The middle peak indicated regions covered only by half of the genomes and present in only one of the haplotypes, and the right-most peak indicated regions, which were present in both haplotypes, and showed the expected coverage. In regions represented in both haplotypes, 578,209 SNPs were defined. Regions without SNP markers were classified into 3253 deletions and 7199 conserved regions (see the “Online methods” section). c SNP phasing along contigs. Genotyping was first performed for each individual nucleus at each SNP marker. As shown, both genotypes (in red and blue) were mixed in the curated but mosaic assembly. After phasing, 8 and 7 nuclei were respectively clustered for genotype A and B, and crossover could be identified. With this, representative markers were imputed at ends of contigs. d. Imputation of markers at deletions by genotyping using normalized read count. Two cases were considered for phasing (and positioning) a deletion marker (in the genetic map). If it was linked with surrounding SNP alleles, it could be phased accordingly; otherwise, comparison of its genotype sequence to genotype sequences of all other markers (including SNP-derived markers at ends of contigs) would be performed to find its value of phase (and positioning). e Linkage group and genetic map construction using the set of imputed markers (SNP-derived markers labeled as 1–8 and deletion markers as p and q). For example, the genotype sequences of 6, 8, and q needed to be flipped (i.e., phase values were 1 - contig phasing). Further ordering of the markers (using JoinMap 4.0) led to linkage group-wise genetic maps