Abstract
High-throughput sequencing (HTS) has revolutionized researchers’ ability to study the human transcriptome, particularly as it relates to cancer. Recently, HTS technology has advanced to the point where now one is able to sequence individual cells (i.e., “single-cell sequencing”). Prior to single-cell sequencing technology, HTS would be completed on RNA extracted from a tissue sample consisting of multiple cell types (i.e., “bulk sequencing”). In this chapter, we review the various bioinformatics and statistical methods used in the processing, quality control, and analysis of bulk and single-cell RNA sequencing methods. Additionally, we discuss how these methods are also being used to study tumor heterogeneity.
Keywords: Tumor heterogeneity, Transcriptomics, Single-cell, High-throughput sequencing, Differential expression, Normalization, Quality control
1. Introduction
Cancer research is becoming more computational focused in which researchers require working knowledge of data science methods to mine and extract new insights from large molecular datasets to derive novel hypotheses. Much of this need stems from the initial sequencing of the human genome to the advent of single-cell sequencing technologies, which has resulted in an explosion in the ability of cancer researchers to generate and acquire high-dimensional data. The most common type of ‘omic data currently generated in the study of cancer biological systems is mRNA gene expression or transcriptomic data.
Uncontrolled cell growth and proliferation is a hallmark feature of cancer. In doing so, genes that regulate cell growth and differentiation are altered, thus leading to the uncontrollable growth and oncogenesis. Often, these changes in gene regulation are caused by mutations that disrupt the “normal” function of the gene and the downstream protein, such as in the case of p53 [1]. In other cases, the gene regulation is changed by aberrant DNA methylation, where by silence the expression of a given gene [2]. Thus, studying the gene expression gives us clues into the biological mechanism of oncogenesis, along with tumor progression and growth. Additionally, gene expression has been used to subclassify a tumor class into smaller, more homogeneous molecular subtypes. For example, gene expression studies have been able to successfully subclassify breast cancer tumors into four to six molecular subtypes (luminal A, luminal B, HER2-enriched, triple-negative/basal-like, normal-like, claudin low) [3–7].
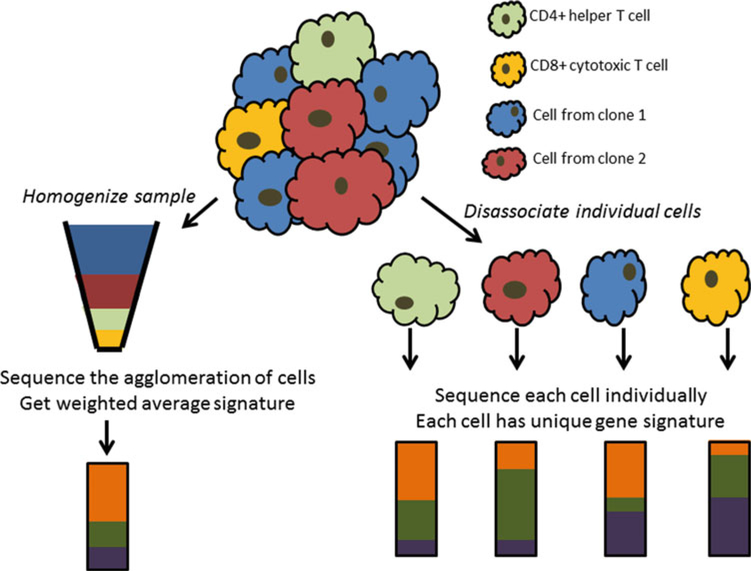
With the advances in technology from microarrays to high-throughput sequencing (HTS), one is able to detect other transcriptomic events, such as gene fusions or chimeras, isoform expression, and allelic expression [8]. Recently, HTS technology has advanced to the point that now one is able to sequence individual cells (i.e., “single-cell sequencing”) [9, 10]. Prior to single-cell sequencing technology, HTS would be completed on RNA extracted from a tissue sample made up of multiple cell types (i.e., “bulk sequencing”). The difference between single cell sequencing and bulk sequencing of RNA is that in the former the sequencing library represents a single cell while the latter represents a population of cells (Fig. 1). Single-cell technology allows researchers study the transcriptome of different cells within the same tissue type. This technology is particular useful in studying cancer immunology and the dissection of tumor heterogeneity, as tumors and the stromal component of tumors are a composition of (1) different cancer cells developed from different genomic events (i.e., clones, tumor heterogeneity) [11, 12] and (2) mixture of cancer cells and immune cells (i.e., tumor infiltrating lymphocytes (TILs)) [13, 14]. In the following sections we will outline the various bioinformatics and statistical methods used in the analysis of bulk and single-cell RNA sequencing.
Fig. 1.
Illustration of differences between bulk RNA sequencing (RNA-seq) vs. single-cell RNA sequencing (scRNA-seq)
2. Datasets Used to Illustrate the Methods
Melanoma is the fifth most common malignance in the United States and it is estimated that 96,480 individuals will be diagnosed with melanoma in 2019 and that an estimated 7230 will die from melanoma [15]. Many genomic studies have been conducted to understand the molecular features of melanoma. Data from The Cancer Genome Atlas (TCGA) determined four major subtypes of cutaneous melanoma: BRAF mutant (52% of tumors), RAS mutant, NF1 mutant, and Triple Wild-Type [16]. It was also found that the immune system plays a central role in the progression and treatment response in melanoma patients. To better understand the influence of immune system and tumor heterogeneity in melanoma, many studies have recently been completed to understand melanoma at the single-cell level [17–20]. To illustrate the bioinformatic and statistical methods used in the analysis of bulk and single-cell RNA-sequencing, we will use data from the TCGA melanoma study [16] and the Tirosh et al. study [20].
2.1. Bulk RNA-Sequencing Study: TCGA Skin Cancer Study
The RNA-seq summarized gene expression levels of skin cutaneous melanoma study (SKCM) using data obtained from TCGA project were downloaded via Genomic Data Commons (GDC) [16]. To illustrate differential expression analysis using RNA-seq data, set out to determine differentially expressed genes between primary tumors (N = 67) and metastatic tumors (N = 213). After filtering nonexpressed or low-expressed genes based on counts per million (CPM), 22,236 genes with CPM values above 1 in at least four libraries remain. To illustrate assessment of batch effects, technical artifacts were downloaded from MBatch (https://bioinformatics.mdanderson.org/tcgabatcheffects) for the SKCM TCGA data. This data set included the following variables: tissue source site (25 levels), plate ID (16 levels), batch ID (14 levels), and ship date (14 levels).
2.2. Single Cell Sequencing Study: Tirosh et al. Study
Tirosh et al. [20] measured single-cell RNA-seq gene expression of 4645 melanoma, immune, and stromal cells from 19 melanoma tumors. These tumors included one primary acral melanoma, ten metastases to lymphoid tissues, and eight metastases to distant sites. The immune (CD45+) and nonimmune (CD45−, including melanoma and stromal) cells were sorted into 96-well plates by flow cytometry (fluorescence-activated cell sorting). Single cell RNA was then isolated and sequenced with SMART-Seq2 protocol [21]. The gene expressions were quantified as y = log2 (TPM + 1), where TPM refers to transcripts per million. Cells with either fewer than 1700 detected genes or average housekeeping gene expression below 3 were excluded.
3. Statistical and Bioinformatics Methods for Analysis of Bulk RNA-Seq Data
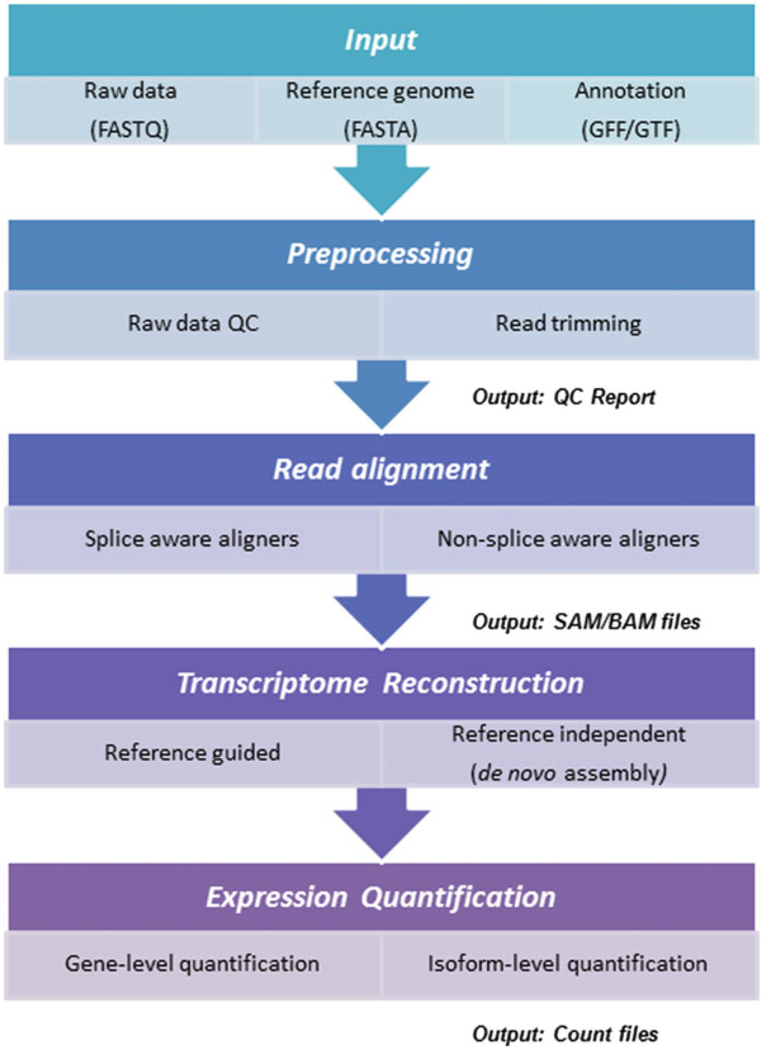
Current RNA-seq protocols still possess several essential biases and limitations, such as nucleotide composition bias, GC content, and polymerase chain reaction artifact or contaminations [22, 23]. Raw RNA-seq data must be checked and processed by quality control (QC) procedures to ensure accurate transcript measurements. Initial steps in the QC process typically involve assessing such biases of the raw reads using metrics generated by the sequencing platform or calculated directly from the raw reads (Table 1). One of the most popular tools for the generation of these quality metrics is FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The RNA-seq raw data and alignment files include various formats such as FASTA to store reference genome [24], gene transfer format (GTF) to store transcript/gene annotations, FASTQ to store raw read data [25], and the sequence alignment map (SAM/BAM) to store read alignments [26]. RNA-seq analysis typically is consisted of major steps including raw data quality control (QC), read alignment, transcriptome reconstruction, expression quantification, and end with downstream analysis (Fig. 2).
Table 1.
Selected list of RNA-seq analysis tools for preprocessing, read alignment, transcriptomic reconstruction, and expression quantification/abundance estimation
| Workflow | Category | Software tools | Reference |
|---|---|---|---|
| Preprocessing | RAW data QC | FastQC | Babraham Bioinformatics website |
| HTQC | Yang et al. [182] | ||
| Read trimming | NGS QC | Patel and Jain [183] | |
| FASTX-Toolkit | Cold Spring Harbor Laboratory website | ||
| Trimmomatic | Bolger et al. [184] | ||
| SolexaQA | Cox et al. [185] | ||
| Read alignment | Spliced aligner | TopHat | Trapnell et al. [186] |
| STAR | Dobin et al. [30] | ||
| MapSplice | Wang et al. [29] | ||
| GSNAP | Wu et al. [31] | ||
| Unspliced aligner | Stampy | Lunter and Goodson [32] | |
| MAQ | Li et al. [33] | ||
| BWA | Li and Durbin [34] | ||
| Bowtie2 | Langmead and Salzberg [35] | ||
| Transcriptome reconstruction | Reference-guided | Cufflinks | Trapnell et al. [36] |
| StringTie | Pertea et al. [37] | ||
| Reference-independent | Trinity | Grabherr et al. [39] | |
| Oases | Schulz et al. [40] | ||
| transABySS | Robertson et al. [187] | ||
| Expression quantification | Gene-level quantification | featureCounts | Liao et al. [43] |
| HTSeq | Anders et al. [44] | ||
| Isoform-level quantification | Cufflinks | Trapnell et al. [36] | |
| StringTie | Pertea et al. [37] | ||
| RSEM | Li and Dewey [41] | ||
| Sailfish | Patro et al. [42] | ||
Fig. 2.
Typical bulk RNA sequencing workflow
3.1. Quality Control of RNA-Seq Data
QC of raw data should be performed as the initial step which involves assessing such biases using metrics generated by the sequencing platform or calculated directly from the raw reads. In addition, depending on the RNA-seq library construction strategy and sequencing [27], trimming strategies include “adapter trimming” and “quality trimming” can be used to remove low-quality reads, trim adaptor sequences, and eliminate poor-quality bases. Adapter trimming is not necessary as most recent sequencers provide raw read in which the adapters are already trimmed, while quality trimming may be an important step depending on the analysis procedure used. Table 1 represents the widely used sequencing QC software tools.
3.2. Methods for Read Alignment
Reference-based alignment is the process used to determine the potential mapping locations by exact match or scoring sequencing similarity. Reads are typically aligned to either a genome or a transcriptome as a reference using two common approaches; (1) splice-aware read aligner or (2) nonsplice-aware read aligners (Table 1). The spliced read aligners use of a gapped or spliced mapper as reads may span splice junctions. Various spliced aligners have been developed including TopHat2 [28], MapSplice [29], STAR [30], and GSNAP [31]. Unspliced read aligners do not allow large gaps, such as those arising from reads spanning exon boundaries, or splice junctions including Stampy [32], mapping and assembly with quality (MAQ) [33], Burrow–Wheeler Aligner (BWA) [34], and Bowtie2 [35].
3.3. Methods for Transcript Reconstruction
Transcript reconstruction includes two approaches to identify all transcripts expressed in a specimen depends on the presence or absence of a reference sequence (Table 1). When the reference annotation information is well-known, then the reference-based approaches such as Cufflinks [36] and StringTie [37] are used to reconstruct transcripts by assembly of overlapping aligned reads. When a reference genome or transcriptome is not available or is incomplete, assembled de novo algorithm directly builds transcripts from short reads using platforms such as Trinity [38], transABySS [39], and Oases [40].
3.4. Methods for Gene Summarization or Abundance Estimation
One of the most widely used applications of RNA-seq is to quantify expression levels of genes and transcripts. Generally, the methods for gene quantification can be divided into two categories: “union exon”-based and “transcript”-based approaches (Table 1). Transcript-based approach fundamentally distributes reads among transcript isoforms including RSEM [41], Cufflinks [36], and StringTie [37]. However, some transcript-based quantification tools such as Sailfish [42] are alignment-free tools to estimate isoform abundances directly from a set of reference sequences. The “union exon”-based methods, such as featureCounts [43] and HTSeq [44], are widely used in RNA-seq gene quantification because of its simplicity to aggregate raw counts of mapped reads.
3.5. Normalization
Variability in measurement can be attributed to both the biological and technical factors. Sources of technical variation, involving, differences in library preparation across samples, sequencing error, mapping and annotation bias, sequencing composition and similarity, gene length, and sequencing depth [45–47] that can significantly reduce the accuracy of statistical inferences and also prevent researchers from properly modeling biological variation and group-specific changes in gene expression [48–51]. Some sources of between-sample technical variation are due to differences in library size or sequencing depth [47]. To correct for library size, most of methods use a common scaling factor per sample to normalize genes such as upper quartile (UQ) [45], median (Med) [45], relative log expression (RLE) [52], trimmed mean of M-values (TMM) [53], and quantile (Q) [54, 55]. Many of these methods are implemented in the edgeR and DESeq2 packages.
Additionally, gene length impacts the comparison of abundance estimates between genes [56], as longer genes contribute more sequenced fragments compared to shorter ones [47]. Most commonly used methods for gene length correction include TPM (transcripts per million) [57] and RPKM/FPKM (reads/fragments per kilobase per million mapped reads) [36, 47], where the former one is considered more robust to differences in RNA library size [58, 59]. However, it has been shown that scaling by gene length cannot entirely remove the positive association between gene size and read counts, and can introduce new biases to the estimates of differential expression [45, 60]. Usual normalization approaches mostly adjust the sequencing depth or/and gene length and fail to correct known or unknown technical artifacts due to the complex. To adjust known batch effect, appropriate statistical models were proposed such as linear regression model or flexible empirical Bayes method (ComBat) which is more robust and appropriate for small sample sizes [61]. ComBat can be implemented in R using the sva package and the function “ComBat.”
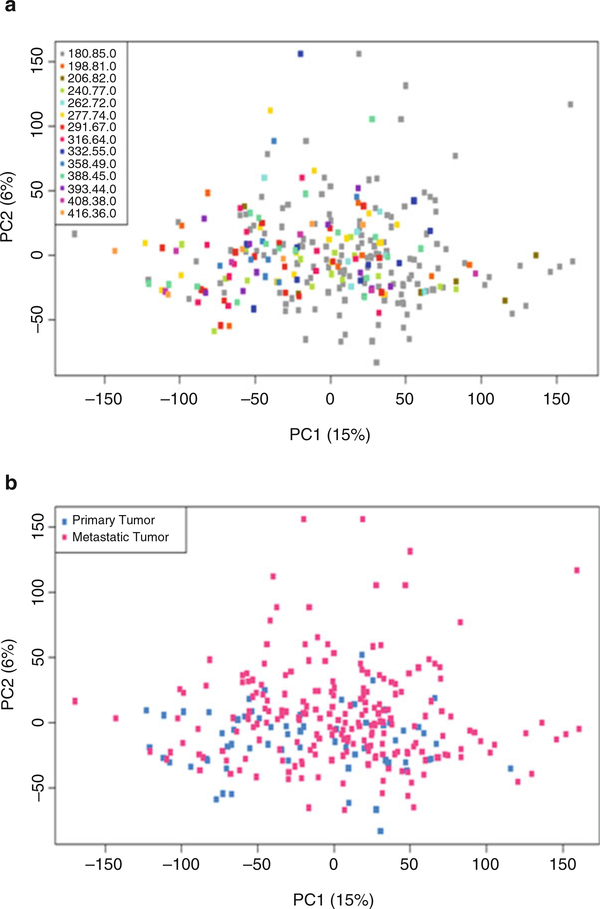
In addition to correcting for known artifacts, assessment and adjustment for potential latent factors is also warranted where mostly rely on singular value decomposition (SVD) or some other factor analysis approaches (e.g., Remove Unwanted Variation (RUV) in R package “ruv” [50] or surrogate variable analysis (SVA) [49] in R package “sva”). Finally, principal component analysis (PCA) is often used in which a subset of the principal components are used to normalize the data [62–65]. Note that the choice of normalization method to remove technical artifacts can affect noticeably the results of differential gene expression analyses [45, 53, 66]. For the TCGA skin cancer study, PCA was completed using filtered raw counts to assess the effects of the batch ID (Fig. 3a) and biological factor (i.e., primary and metastatic tumors groups) (Fig. 3b). Figure 3a represents the effect of batch ID with high proportion of variation for top principal component (15%) which leads to consideration of batch ID as a technical effect for normalization.
Fig. 3.
Plots from Principal Component Analysis to assess technical batch and biological factor effects globally in bulk RNA-seq experiments for the TCGA skin cancer study. (a) Fourteen levels of known batch ID, where batch ID was downloaded from https://bioinformatics.mdanderson.org/BatchEffectsViewer/; and (b) primary factor of interest (primary tumor and metastatic tumor) using filtered raw counts data
3.6. Methods for Differential Gene Expression Analysis
The main methodologies for differential gene expression analysis for RNA-seq data are categorized by the distributional assumptions (Table 2). Models for read counts originated from the idea that the number of reads for each gene can be approximated by a Poisson distribution where log-linear or generalized linear model were proposed to model the mean difference between samples along with using test statistics such as likelihood ratio test, exact test, and score test for hypothesis testing are implemented in DEGseq [67], Myrna [68], and PoissonSeq [69] packages. The extended Poisson models, two-stage Poisson model [70] and generalized Poisson model [71] are also considered to adjust for overdispersion issue.
Table 2.
Statistical methods to identify differential gene expressions based on RNA-seq data
| Model | Software | Reference |
|---|---|---|
| Poisson | DEGseq | Wang et al. [67] |
| Myrna | Langmead et al. [68] | |
| PoissonSeq | Li et al. [69] | |
| Negative binomial | edgeR | Robinson et al. [73] |
| DESeq | Anders and Huber [52] | |
| DESeq2 | Love et al. [74] | |
| NBPSeq | Di et al. [75] | |
| Beta-binomial | BBSeq | Zhou et al. [76] |
| Bayesian and empirical Bayesian | ShrinkSeq | Van de Wiel et al. [77] |
| baySeq | Hardcastle and Kelly [78] | |
| Normal | limma+voom | Smyth [54, 79] |
| Law et al. [81] | ||
| Nonparametric | SAMseq (samr) | Li and Tibshirani [82] |
| NOIseq | Tarazona et al. [83] | |
Poisson and negative binomial distributions are the two widely used models [52, 72], whereas the higher variability between biological replicates leads to incorporate negative binomial distribution to accommodate overdispersion [45]. The dispersion parameter estimation can be based on the conditional maximum likelihood, pseudo-likelihood, quasi-likelihood, local regression, and conditional inference using hypothesis testing approaches such as Wald test, likelihood ratio test, and exact test. Such methods are included in edgeR [73], DESeq [52], DESeq2 [74], and NBPSeq [75] packages. A beta-binomial model is implemented in BBSeq [76] package which accommodates the overdispersion using logistic regression where maximum likelihood approach is applied to estimate overdispersion parameter. Moreover, full or empirical Bayesian frameworks are included in ShrinkSeq [77] and baySeq [78] packages. Lastly, methods have been proposed that allow RNA-seq to be modeled using a linear model framework (i.e., Gaussian distribution) through limma package [79, 80] using voom transformation [81] or approaches that do not assume any distribution assumption (nonparametric approaches) such as SAMseq [82] and NOIseq [83]. It should be noted that if the modeling assumptions are valid the parametric methods will be more powerful than the nonparametric methods. However, if the modeling assumptions are not valid, nonparametric methods would be advisable. In practice, it is difficult to assess the modeling assumptions and thus parametric methods are mostly often used, particularly when the sample size is small. In skin-TCGA study, to identify the differentially expressed genes between primary and metastatic tumors groups (Fig. 3b), the voom-transformed UQ normalized data is considered where the design matrix contains the estimated latent artifact and the batch ID along with the tumor groups using limma package.
3.7. Methods for Correcting for Multiple Testing
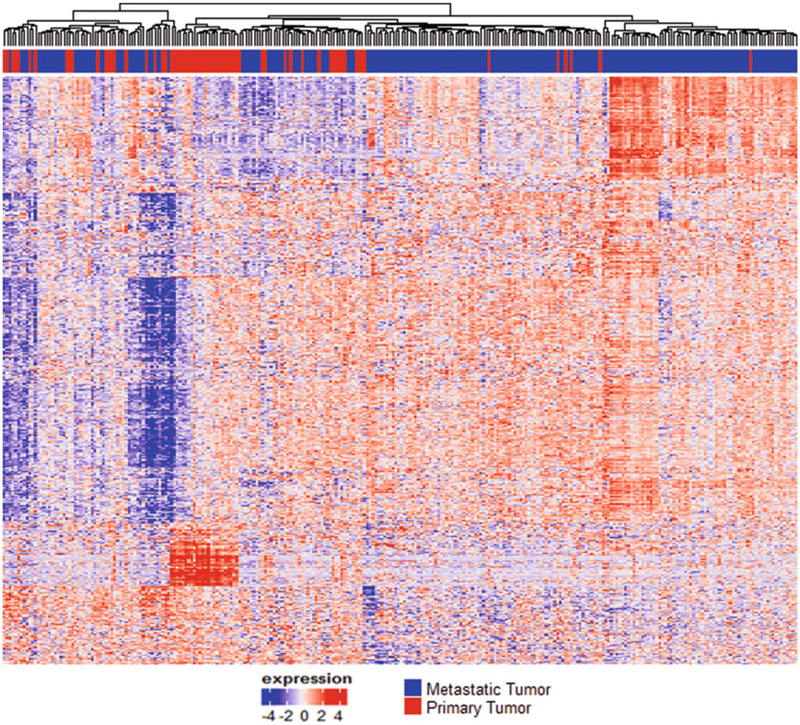
With thousands of genes to test, controlling both the overall Type I error rate and the desired statistical power becomes important. Multiple comparison adjustment approaches can control the familywise error rate (FWER), false discovery rate (FDR) (i.e., Benjamini and Hochberg approach [84]), and Bayesian FDR (q-values) [85, 86]. Various approaches control FWER and compute the adjusted P values such as Bonferroni [87], Holm [88], and Hochberg [89]. In contrast to the strong control of FWER, the FDR-based control is less conservative with the increased gain in power and has been widely used in cases where a large number of hypotheses are simultaneously tested. Figure 4 represents the heatmap of top differentially expressed genes (n = 8928) after correcting multiple testing, FDRBH < 0.05.
Fig. 4.
Heatmap of the top differentially expressed genes (FDRBH < 0.05) from the analysis of the TCGA skin cancer study
3.8. Studying TME Using RNA-Seq in Bulk Samples
A software tool CIBERSORT is widely used to estimate fractions of multiple cell types using gene expression data in bulk samples [90]. It is commonly used to characterize global immune landscape by estimating different proportions of different immune cells. For instance, in a recent large-scale study to characterize immune landscape by analyzing 10,000 tumors comprising 33 diverse cancer types, CIBERSORT was used to estimate immune infiltration fractions for understanding tumor–immune interaction [91]. Six immune subtypes identified are wound healing, IFN-g dominant, inflammatory, lymphocyte depleted, immunologically quiet, and TGF-β dominant across cancer types and provided this as a source, iAtlas (https://www.cri-iatlas.org/), for researchers to understand tumor–immune interaction and potential therapeutic opportunities. In addition to cell type identification, the cell–cell interaction from the ligand and receptor database is also incorporated. Although CIBERSORT is widely used, its performance potential is affected by statistical multicollinearity due to the inclusion of highly correlated immune cell types, and also was developed using expression on microarrays. TIMER is developed to select genes, which are negatively correlated with tumor purity for each cancer type, and then apply constrained least squares fitting to expression to predict the abundance of a subset of TILs: B cells, CD4 T cells, CD8 T cells, macrophages, neutrophils, and dendritic cells [92]. To capture the complexity in TME better, xCell attempted to infer 64 immune and stromal cell types by using gene set enrichment analyses and deconvolution method to analyze the 1822 harmonized human pure cell type transcriptomics samples [93].
4. Statistical and Bioinformatics Methods for Analysis of Single-Cell RNA-Seq Data
Single-cell RNA-sequencing (scRNA-seq) has been playing important roles in the study of tumor heterogeneity and tumor evolution. In contrast to the bulk RNA-seq where the average gene expressions are measured across a large population of cells, scRNA-seq quantifies transcriptome of individual cells. With the newly developed high-throughput cell separation technologies, thousands of cells per tumor can be profiled in parallel to capture intra-tumor heterogeneity at an unprecedented resolution. Multiple different platforms have been developed for scRNA-seq including SMART-seq [21], CEL-seq [94], Fluidigm C1 [95], Smart-seq2 [96], and more advanced droplet-based platforms including Drop-seq [97] and Chromium 10X [98]. In droplet-based platforms, cells are encapsulated in water-based droplets together with unique molecular identifiers (UMIs), a cell-specific and transcripts-specific barcoding system. These barcodes help to diminish the sequencing reads representation biases due to library amplification.
The considerable differences in cell isolation and molecule capture lead to large variations in sensitivity, specificity, and capacity of these platforms [99, 100]. However, they all rely on similar computational pipelines to reveal transcription dynamics. In the following sections, we will review the algorithms in major steps of scRNA-seq data analysis using the most commonly used droplet-based platforms, but the discussion applies to all platforms.
4.1. Quality Control
The droplet-based scRNA-seq platforms encapsulate thousands of cells individually into barcoded-droplets and sequence their RNA material simultaneously. All computational analyses and interpretations of results reply on the assumption of single-cell behavior, such that only one living cell exists in a single droplet. However, even for the most sensitive protocols, it is inevitable to have dead cells and doublets (multiple cells encapsulated in one droplet) [101]. Therefore, it is essential to apply quality control (QC) to identify the low-quality droplets/barcodes/cells which ought to be excluded from downstream analyses [97, 101–105].
A common QC metric for scRNA-seq is the number of transcripts/UMIs detected per droplet. A small number of transcripts detected per barcode are often an indicator for poor droplet capture, which can be caused by cell death and/or capture of random floating RNA molecules released by dead cells. Inversely, a considerable large number of transcripts with the same barcode can suggest doublets or floating RNA encapsulated together with a living cell. Percentage of mitochondrial transcripts is another common QC metric. A high number of mitochondrial transcripts suggest the cells might be undergoing stress, for example, from cell isolation and sorting process. It is advised to remove these cells since stress level is usually not the interests of scRNA-seq analysis. In addition to the cell-level QC, gene-level QC is often performed to exclude genes expressed in only a very small proportion of cells. Removing such genes can decrease technical noise, and speed up the downstream normalization and clustering.
When deciding on the cutoffs for scRNA-seq QC, it is important to take into the consideration the cell compositions of the samples being analyzed. Different cell types actively express different number of genes, different number of mitochondrial transcripts, and different sets of transcripts, especially when comparing tumor to normal cells. A too stringent cutoff can remove a cell population of interests if this population is rare in sample of cells. Therefore, we suggest that if the researchers have no prior knowledge about the cell compositions, a possible solution is to carry out initial cell type identification and use that information to guide the QC process.
4.2. Drop-Outs, Normalization, and Spike-Ins
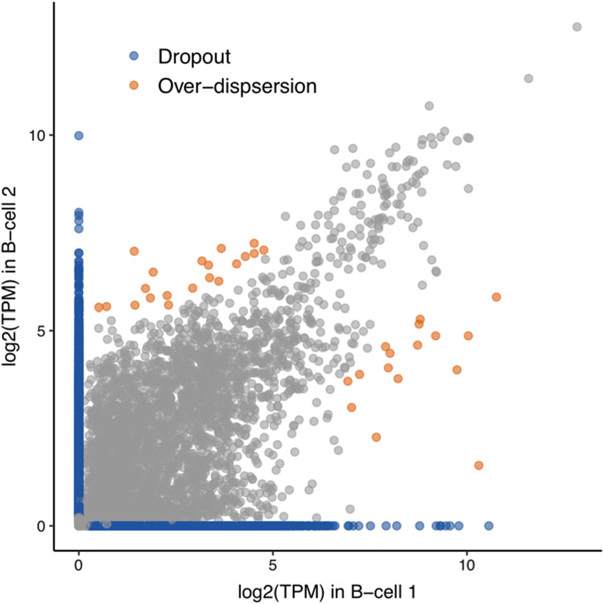
When analyzing scRNA-Seq data, normalization is a critical step to adjust for unwanted biological effects and technical noise collectively known as “batch effects” that mask the real signal. Similar to bulk RNA-seq, scRNA-seq batch effects can come from the variations in handling protocols, library preparation, sequencing platforms, and sequencing depth. In addition to these variations commonly seen in bulk RNA-seq, a prominent characteristic of scRNA-seq data is zero inflation, where the expression count matrix of single cells is mostly filed with zeros [106–108] (Fig. 5). There are two sources of zero inflation: (1) biological reason—the real zeros, where the cells are in transient state of transcript bursting [109] or the genes simply do not express in a subpopulation of cells; (2) technical reason—dropout, which is caused by the inefficiency of mRNA capture such that a large percentage of mRNA molecules are not captured and consequently not sequenced. Besides the dropout events, individual cells can show stronger overdispersion than typically observed in bulk RNA-seq data, even for genes with median-to-high expression levels [108]. Furthermore, compared to bulk RNA-seq, scRNA-seq data is much more heterogeneous since the sequenced cells are usually of different populations, cell types, or statuses. Even the cells from the same population but undergoing different cell cycles can show very different expression profiles [110, 111]. To uncover the cellular heterogeneity, scRNA-seq studies often start with choosing the HVG (highly variable genes) that are most informative in distinguishing cell populations. It has been shown that the choice of HVG is highly affected by normalization [112, 113]. Therefore, it is important for scRNA-seq normalization to retain the cell-to-cell biological heterogeneity while removing the cell-specific noise at the same time.
Fig. 5.
Cell-to-cell variability observed in scRNA-seq data of two single B cells
4.2.1. Normalization Methods
The global-scaling normalization methods inherited from bulk RNA-seq analysis have been widely used in scRNA-seq. However, these methods cannot accommodate the cell-specific variability in scRNA-seq data and can lead to biased estimation of scaling factors [113–115]. To optimize the modeling of the cell-to-cell variability, multiple normalization methods specifically tailored for scRNA-seq data have been developed. Table 3 provides a detailed summary of their main features, statistical models, and special considerations when modeling cell-specific biological and technical variations. In the following sections, we will briefly discuss the key characteristics of these methods.
Table 3.
Normalization methods used in scRNA-seq data analysis
| Method | Features | Statistical models | Notes on biological variation | Notes on technical variation | Other factors |
|---|---|---|---|---|---|
| SCDE [108] | A two-component mixture model is used to capture dropout events and amplification events; Differential expression is evaluated by Bayesian approach | 1. Dropouts: Poisson 2. Amplification: Negative binomial |
|||
| TASC [116] | An empirical Bayes approach models the cell-specific dropout rates and amplification bias | 1. Biological variation: Poisson 2. Technical variation: Logistic regression |
Cell-specific technical variation is modeled by spikes-ins | Cell cycle and cell size are considered as covariates | |
| MAST [106] | Two-part generalized linear regression models dropout and amplification events | 1. Probability of detection: logistic regression 2. Expression level: Gaussian |
Fraction of genes detected in each cell (CDR) is used as proxy for both biological and technical variation | Cell size is captured by CDR | |
| BASiCS [117] | Use integrated Bayesian hierarchical model to simultaneously quantify unexplained technical noise and cell-to-cell biological heterogeneity | 1. Expression: Poisson 2. Random effect and size-specific factor: Gamma |
Biological cell-specific variability is quantified by gene-specific parameters borrowing information across all cells | Technical variability is quantified based on spike-ins | Cell cycle and cell size are captured |
| SCALE [109] | Allele-specific transcription expression is modeled by a systematic statistical framework | 1. Alleles expression status: empirical Bayes method 2. Allele-specific transcription kinetics: Poisson-Beta hierarchical model 3. Allelic difference: resampling-based test |
Requires allele-specific read counts to start with | Technical variability is adjusted through spike-ins | Spike-ins/total reads servers as a proxy for cell size |
| scran [107] | Deconvolution frameworks estimates cell-specific factor by pooling multiple cells to deal with zero inflation | A ring arrangement by library size and sliding window is used to select random pool of cells; pool-based scaling factors are then deconvolved to yield cell-specific factors | |||
| SCnorm [115] | Use quantile regression to estimate gene-specific scaling factors based on sequencing depth | Quantile regression | Groups genes with similar on sequencing depth and then estimate scale factors within each group; Spiked-ins are not required, but can be helpful |
||
| BISCUIT [118] | Employs Bayesian inference to cluster single cells considering both biological and technical variation in parallel | Hierarchical Dirichlet Process mixture model (DPMM) | The model simultaneously estimates the heterogeneous clusters through the DPMM and infers the technical variation parameters for imputing dropouts | ||
The first category we consider detects differential expressions among cells while adjusting technical variations that are specific to scRNA-seq data. This category includes SCDE [108], TASC [116], and MAST [106], which all employs empirical Bayesian frameworks to estimate dropout events and real amplification of transcripts. More specifically, TASC takes the cell size and cell cycle as covariates, where the former is estimated from the ratio of endogenous RNA reads to spike-ins and the latter is represented by the expression of curated genes [20]. Alternatively, MAST uses a fraction of genes detected in each cell as a proxy for technical and biological variation. Although most of these methods focus on the differential expression at the gene level, some go beyond and study the difference in allelic expression. One such example is SCALE, which models the allele-specific transcription bursting with both technical variation and cell size differences accounted for. However, SCALE requires input as allele-specific read counts at heterozygous loci, which can be challenging for many scRNA-seq platforms, especially the tag-based quantification methods including Drop-seq and 10X.
Another major category is to generate normalized gene expression matrix that can be used as input for downstream analysis. Based on how the scaling factors are modeled, we further stratify these methods into two groups: cell-specific methods including scran [107] and BASiCS [ 117], and gene-specific methods including SCnorm [115]. BASiCS estimates the cell-specific biological variations by borrowing information across all cells and all genes while quantifies the technical variations replying on spike-ins. Alternatively, scran first clusters the cells into more homogenous groups and then deconvolutes the pooled cells to yield cell-specific factors. On the other hand, SCnorm groups genes with a similar dependence on sequencing depth and then estimates the scale factor within each gene group.
In addition to above approaches that focus on differential expression and normalized gene expression matrix, which are concepts adopted from bulk RNA-seq, several methods have been developed to specifically target the downstream heterogeneity studies in scRNA-seq data. BISCUIT [118] is a cell-type dependent normalization which uses a Bayesian probabilistic model to iteratively normalize and cluster cells. It simultaneously assigns cells to cluster and learns cell-dependent parameters within each cluster. The inferred parameters are then used to generate cell-type dependent normalization that can be fed back to improve clustering. It has been shown that BISCUIT can identify more refined subtypes of cells than global normalization methods [119].
4.2.2. Drop-Out Imputation
Instead of directly normalizing endogenous genes, many chose to impute the drop-out events prior to normalization. There are mainly two strategies used in scRNA-seq data imputation:
To distinguish the biological zeros from technical zeros using models of expected gene expression, which is usually obtained either from borrowing information across cells or from spike-in sequences. These methods include DrImpute [120], SAVER [121], McImpute [122], scImpute [123], ALRA [124], and scRMA [125].
To reduce the noise by using information from neighboring data. This category includes MAGIC [126], netSmooth [127], and knn-smooth [128].
Although imputation can rescue missing information that is important to study cellular heterogeneity, concerns have been raised about their sensitivity and specificity. A positive-control based benchmarking study assessed these methods and concluded that most methods only provide small improvement [129]. Andrews and Hemberg [130] evaluatedthe false discovery rate of these methods using negative controls. They found that SAVER performed well on simulated data compared to others; however, all methods introduced false signals at various levels in the permuted real data. These limitations of imputing scRNA-seq data are probably due to the lack of a comprehensive and independent reference, for example, as in GWAS imputation. Until such reference is generated, caution should be used when imputing scRNA-seq data.
4.2.3. Spike-Ins
As shown in Table 3, many scRNA-seq normalization methods rely on the spike-ins to estimate the cell-specific technical variations. Spikes-ins are a set of synthetic RNA sequences added to the samples in a theoretically constant and known amount, in order to calibrate the gene measurement and distinguish the biological vs. technical variations in RNA-seq experiments. The most commonly used spike-ins are the 92 External RNA Control Consortium (ERCC) molecules [46] and the Spike-in RNA variant control mixes (SIRVs, Lexogen). These extrinsic control sequences have also been used in scRNA-seq experiments [131–133], where the spike-ins with different concentrations are added with a constant amount across all cells. Vallejos et al. [113] and Lun et al. [134] have discussed the benefit of extrinsic control sequencing in scRNA-seq. However, the use of spike-ins remains challenging.
Although often neglected, calibrating the amount of spike-ins sequences is critical and should depend on the endogenous mRNA content [50]. However, due to the large and unknown heterogenous among tumor microenvironment, it is difficult to obtain prior knowledge about the cell-type specific endogenous mRNA content before sequencing. In addition, the spike-in sequences do not reflect the gene-length and GC content in the mammalian transcriptome, such that the technical effects may be different for the extrinsic and intrinsic genes [113]. Moreover, it has been shown that spike-ins signals can vary across technical replicates [50], and only partial spike-in sequences can be actually sequenced and aligned [113]. Furthermore, the use of spike-ins in the recent developed large-scale droplet-based platforms is not as cost-effective as in small scale platforms. For example, to reduce the doublet rate in Chromium 10X, the percentage of cell-containing droplets is deliberately designed as low as 1–10%. The spike-ins are added evenly across all droplets, not just the cell-containing ones and consequently takes up the vast majority of sequencing reads. Due to these limitations and challenges, caution should be used when employing spike-ins for technical variation estimations. Additionally, efforts should be made to design spike-ins sequences accommodating the unique characteristics of scRNA-seq experiments.
As discussed above, normalization methods that are specially tailored for scRNA-seq are theoretically and operationally superior over the global-scaling normalization inherited from bulk RNA-seq. However, these methods vary substantially in terms of their assumptions and their models, where none of them outperform others under all scenarios in the performance assessment [114, 135]. Great efforts are actively underway to develop more efficient and robust normalization for scRNA-seq.
4.3. Data Integration and Batch Correction
Due to the complexity of scRNA-seq experiments, it is often difficult for a study to process all samples at the same time and/or using the same protocols. In such situations, it is necessary to integrate samples of different batches or even of different scRNA-seq platforms. Due to the unique data structure of scRNA-seq, batch correction methods designed for bulk RNA-seq are not suitable. Several approaches have been recently proposed to deal with sample-level batches in scRNA-seq data. kBET (k-nearest neighbor correct effect) [136] quantifies the sample-level batch effects using a χ2-based test. MNNs (mutual nearest neighbor) [137] corrects the effect using only a subset of populations shared between batches and has been implemented in scran [102] as the mnnCorrent function. The Seurat group [138] uses cell pairwise correspondences between single cells across datasets, termed as “anchors,” to integrate gene expressions across technologies and batches. All these approaches have been shown to have better performance than bulk RNA-seq batch-correction methods.
4.4. Dimension Reduction, Clustering, and Cell Type Identification
One of the most popular uses of scRNA-seq is to identify and characterize cell types within the heterogeneous tissues or samples. The de novo identification of putative cell types has been considered as an unsupervised clustering problem. In this section, we will discuss the main classes of clustering methods having been applied to scRNA-seq, as well as the remaining issues and challenges. We will also briefly touch on the supervised and semisupervised clustering.
4.4.1. Dimension Reduction and Feature Selection
The high dimensional transcriptome data generated by scRNA-seq provides tremendous information for uncovering the biology of cells. However, it also introduces challenges to statistical analysis which is often referred to as the “curse of dimensionality.” With a large number of genes measured in scRNA-seq, the distance between individual cells can become small and thus make it difficult to distinguish between-population differences and within-population differences [139]. The two main approaches to deal with the issue of high dimensionality are feature selection and dimension reduction.
Feature selection removes the uninformative genes in terms of their ability to distinguish cells, in order to reduce the dimensions used in analysis and speed up calculations. The most commonly used feature selection in scRNA-seq is to select the highly variable genes (HVG), assuming that genes with high variance are more likely due to biological signals rather than technical noise [140]. The normalization and usage of spike-ins can facilitate the selection of HVG as we discussed in the previous section. Other feature selection approaches include identifying biological relevant genes based on expression correlation between cells [97, 141, 142] and using the magnitude and/or significance of the correlation to select genes.
Dimension reduction, on the other hand, completes a projection of the gene expression data onto a lower dimensional space. There are many generic dimension reduction methods that can be applied to any high dimensional data, including principal component analysis (PCA), independent components analysis (ICA) [143], Laplacian eigenmaps [144], and t-distributed stochastic neighbor embedding (t-SNE) [97, 145]. In this section we will focus on PCA and t-SNE due to their popularity in scRNA-seq data analysis. PCA uses the orthogonal transformations to project the gene count matrix onto a reduced number of linear independent dimensions called principal components. The advantage of PCA is that it is relatively fast and can preserve the distance information among cells. However, PCA is restricted to linear combinations of variables, which can be inappropriate in the context of scRNA-seq. In particular, it has been reported that the first components are often related to the number of genes detected per cell rather than the biological signals [106, 146]. To improve PCA, Risso et al. [147] proposed ZINB-WaVE which uses a zero-inflated negative binomial to deal with the dropouts in scRNA-seq data.
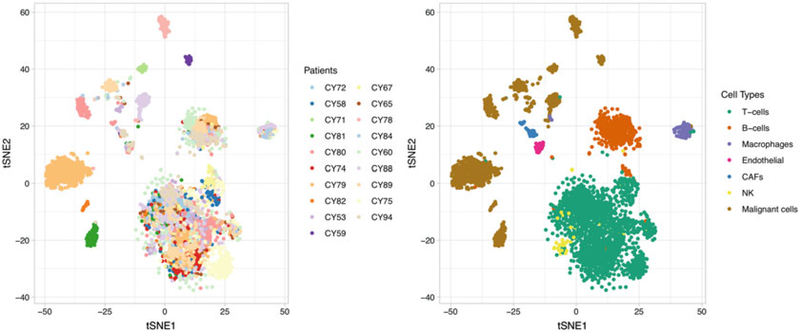
T-distributed stochastic neighbor embedding (t-SNE) is a stochastic method that reduces high dimensions to two or three embeddings while preserving the local structure among cells, such that neighbor cells stay close and distant cells remain distant. Due to the probability distribution used in embedding estimation, t-SNE follows two rules: (1) all points (cells) repel each other, and (2) each point (cell) attracts its nearest neighbors [148]. Therefore, t-SNE can specifically project cells into more distantly isolated clusters, making it almost the standard choice in visualization exploration of scRNA-seq data. Figure 6 projects 4645 single cells extracted from 19 melanoma tumors [20] onto t-SNE 2D planes. The clusters formed by t-SNE are in good agreement with the cell types identified by the original study, and show a high degree of intra-tumor heterogeneity for malignant cells but not for immune/ stromal cells. However, t-SNE has its limitations. First, t-SNE is computationally expensive. This is particularly problematic in large-scale scRNA-seq studies which require analyzing hundreds of thousands of cells simultaneously. In addition, although t-SNE captures the local structure it often fails to preserve the global geometry of the data. When t-SNE places cells into distinctive clusters, the relative position of these clusters is almost arbitrary and with little biological meanings [148]. Moreover, the embedding is governed by a parameter “perplexity,” which controls the number of nearest neighbors each point is attracted to. Different perplexity choices can lead to different degrees of separation and the judgment of the appropriate perplexity is subject to the analysts. Several reviews have provided in-depth discussion and practical suggestions for using t-SNE in scRNA-seq analysis [148–150]. We would recommend readers consult with these reviews before making conclusions with t-SNE results.
Fig. 6.
t-SNE projection of single cells from melanoma tumors colored by patient origin (left) and cell type (right)
More recently, a nonlinear dimension reduction method, uniform manifold approximation and projection (UMAP) [151], implemented in R package “umap,” was developed as an alternative to t-SNE. It is claimed to preserve as much of the local structure and more of the global geometry with a shorter run time than t-SNE. Becht et al. [152] performed a systematic evaluation using well-annotated scRNA-seq data, and concluded that UMAP provided faster run times, higher reproducibility, and more meaningful cell clusters. Recognizing its advantage, several scRNA-seq analysis tools incorporate UMAP into their standard dimension reduction and visualization pipeline. Feature selection and dimension reduction are not necessarily mutually exclusive. As a matter of fact, dimension reductions are susceptible to the batch effects caused by cell-specific technical variations [106, 153]. Performing feature selection that removes genes with little biological relevant prior to dimension reduction can greatly reduce the technical noise [154]. The combination of these methods has been widely adopted by scRNA-seq analysis pipelines.
4.4.2. Unsupervised Clustering
Recent single-cell sequencing technologies enable the study of tumor or TME heterogeneity at a single cell level [155, 156]. One of the ongoing challenges on scRNA-seq data analyses is that cell type recognition or subpopulation classification since the tumor–stromal interaction has been shown to be important. The diverse cell types would often be visualized in t-SNE space (Fig. 6) or other similar linear or nonlinear projections. Unsupervised clustering is a central component of scRNA-seq analysis, as it can identify cell populations and thus strongly affects any downstream analysis. Although many methods have been developed and applied to scRNA-seq, clustering and the interpretation of clusters are still facing great biological and computational challenges. Kiselev et al. [139] and Andrews and Hemberg [ 154] reviewed the commonly used clustering methods in the context of scRNA-seq and summarized their advantages and limitations. Duo et al. [157] systematically evaluated 15 clustering methods using simulated data and found substantial differences in their performances. In this section, we briefly review the four main classes of clustering methods.
K-means clustering iteratively assigns cells to the nearest cluster center and recomputes the new center. Although this method is fast, it requires a predetermined number of clusters and assumes the clusters are of equal sizes, which can be easily violated in TME studies. Tools that implement K-means include SC3 [158], SIMLR [159], RaceID [160], and pcaReduce [161]. The next method is hierarchical clustering which sequentially merges cells into larger clusters (bottom-up) or divides clusters into smaller communities (top-down). This method is deterministic but more computational expensive than k-means. Many scRNA-seq tools have modified hierarchical clustering either to accommodate low-depth samples by adding imputation of zeros [162] or to improve identification of small clusters by iteratively performing dimension reduction [161, 163]. Hierarchical clustering-based tools include CIDR [162], BackSPIN [163], pcaReduce [161], SINCERA [164], mpath [165], and ascend [166].
The third type of clustering methods is density-based, which can identify dense clusters without any assumption on the shape or size on the clusters. However, it assumes equal homozygosity (density) of the clusters, requires a predetermined density cutoff, and works better with datasets with a large number of cells (e.g., droplet-based scRNA-seq assays). Such methods include DBSCAN [167], GiniClust [168] which employs DBSCAN, and monocle [169]. Lastly, there is graph-based clustering, which identifies cells densely connected by edges. Compared to k-means and hierarchical clustering, graph-based clustering does not require any predefined parameters, makes the minimal assumption on cell populations, and can scale to a large number of cells [170, 171]. The most population application of graph-based clustering combines k-nearest-neighbor graphs and Louvain community detection [171, 172]. However, the main drawback of graph-based clustering is that it heavily relies on how well the scRNA-seq is translated into graph space. Therefore, it is often necessary to perform dimension reduction or feature selection beforehand to boost the search for nearest neighbors. Graph-based clustering has been implemented in multiple tools including Seurat [97], Pheno-Graph [173], densityCut [174], SNN-Clip [175], SACNPY [176], and MetaCell [177].
4.4.3. Supervised Classifier and Cell Type Identification
To characterize the sample heterogeneity, the groups of cells defined by unsupervised clustering are often assigned to different cell types based on enriched canonical markers. In cancer researches, the single-cell type identification has been focusing on the subpopulations of immune cells, stromal cells, and tumor-related cells present in TME and/or the circulating system. However, this canonical workflow has its limitations. First, as summarized above, each clustering method has its own drawbacks and different similarity metrics usually result in different cluster separations. Second, this process relies on the researchers’ knowledge on the signature genes, and it can become arbitrary when making conclusions based on only a handful of genes. Only very recently, alternative methods have been proposed. SuperCT [178] is a supervised classifier (SC)-based machine learning method. It trains a nonlinear SC from bulk and single-cell RNA-seq data of pure cell types covering a wide range of immune and stromal cells, and then uses the trained classifiers to reveal cell types of any scRNA-seq data provided as new input. Another single-cell identifier is SingleR [179]. It constructs a reference database by collecting bulk RNA-seq data from over 1000 samples with pure cell types, and then determines the type of a single cell in scRNA-seq experiment by its Spearman correlation with each sample in the reference database. Although these methods are still immature for application in cancer research due to the limited cancer-specific reference database, they defiantly opened up new avenues for cell type classification in scRNA-seq.
4.5. Studying Heterogeneity Using scRNA-Seq
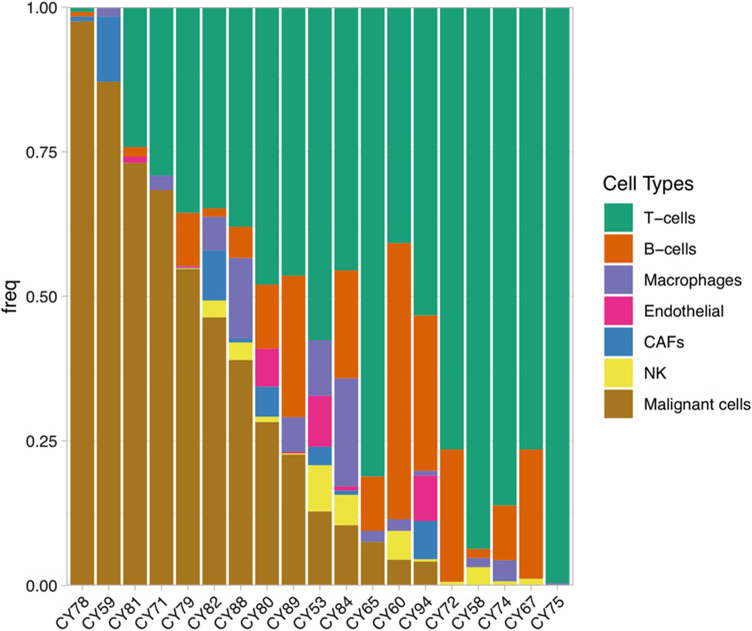
Tumor heterogeneity is commonly observed with wide range of infiltrations, as illustrated in Fig. 7 for 19 melanoma tumors from the Tirosh et al. study [20]. As the algorithms for cell type or subtype classification has been developed and improved in recent years. Some algorithms have been focused on how to quantify cellular heterogeneity. SinCHet estimates cellular heterogeneity using Shannon index over the all-possible clustering resolutions is developed to analyze cellular heterogeneity and characterize subpopulation composition [180]. A recent paper proposes a general diversity index (GDI), a generalized form of ecological diversity index, to quantify heterogeneity on multiple scales and relate it to disease evolution [181]. The index takes the generalized from the low diversity order, the clonal richness, to intermediate diversity, Shannon or Simpson’s indices, to higher order of diversity. The results showed that healthy individuals had lower diversity than cancer patients and little difference in diversity between pre– and post–bone marrow samples from AML patients.
Fig. 7.
Cellular composition of 19 melanoma tumors showing tumor heterogeneity
5. Conclusions
High-throughput sequencing (HTS) has revolutionized the study of the transcriptome and its relationship with disease. Two types of transcriptomic studies are now possible, bulk or single cell sequencing studies. With the advances in technology, many bioinformatics and statistical methods have been developed to process and analyses data from bulk sequencing (RNA-seq) and single-cell sequencing (scRNA-seq), with more methods currently being developed for scRNA-seq studies.
References
- 1.Muller PA, Vousden KH (2013) p53 mutations in cancer. Nat Cell Biol 15(1):2–8. 10.1038/ncb2641 [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB (2005) DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2 (Suppl 1):S4–S11. 10.1038/ncponc0354 [DOI] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB et al. (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752. 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker JS, Mullins M, Cheang MC et al. (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27(8):1160–1167. 10.1200/JCO.2008.18.1370. JCO.2008.18.1370 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorlie T, Perou CM, Tibshirani R et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98 (19):10869–10874. 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorlie T, Tibshirani R, Parker J et al. (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 100 (14):8418–8423. 10.1073/pnas.0932692100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev 10(1):57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu S, Qing T, Zheng Y et al. (2017) Advances in single-cell RNA sequencing and its applications in cancer research. Oncotarget 8(32):53763–53779. 10.18632/oncotarget.17893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bian S, Hou Y, Zhou X et al. (2018) Single-cell multiomics sequencing and analyses of human colorectal cancer. Science 362 (6418):1060–1063. 10.1126/science.aao3791 [DOI] [PubMed] [Google Scholar]
- 11.Navin NE (2015) Delineating cancer evolution with single-cell sequencing. Sci Transl Med 7(296):296fs229 10.1126/scitranslmed.aac8319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee MC, Lopez-Diaz FJ, Khan SY et al. (2014) Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc Natl Acad Sci U S A 111(44):E4726–E4735. 10.1073/pnas.1404656111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo X, Zhang Y, Zheng L et al. (2018) Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 24(7):978–985. 10.1038/s41591-018-0045-3 [DOI] [PubMed] [Google Scholar]
- 14.Zheng C, Zheng L, Yoo JK et al. (2017) Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169 (7):1342–1356.e1316. 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]
- 15.Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69 (1):7–34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Network (2015) Genomic classification of cutaneous melanoma. Cell 161(7):1681–1696. 10.1016/j.cell.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nirschl CJ, Suarez-Farinas M, Izar B et al. (2017) IFNgamma-dependent tissue-immune homeostasis is co-opted in the tumor microenvironment. Cell 170 (1):127–141.e115. 10.1016/j.cell.2017.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerber T, Willscher E, Loeffler-Wirth H et al. (2017) Mapping heterogeneity in patient-derived melanoma cultures by single-cell RNA-seq. Oncotarget 8(1):846–862. 10.18632/oncotarget.13666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar MP, Du J, Lagoudas G et al. (2018) Analysis of single-cell RNA-Seq identifies cell-cell communication associated with tumor characteristics. Cell Rep 25(6):1458–1468. e1454. 10.1016/j.celrep.2018.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tirosh I, Izar B, Prakadan SM et al. (2016) Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352(6282):189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picelli S, Bjorklund AK, Faridani OR et al. (2013) Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10(11):1096–1098. 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- 22.Hansen KD, Brenner SE, Dudoit S (2010) Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res 38(12):e131 10.1093/nar/gkq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y, Speed TP (2012) Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res 40 (10):e72 10.1093/nar/gks001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A 85(8):2444–2448. 10.1073/pnas.85.8.2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cock PJ, Fields CJ, Goto N et al. (2010) The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res 38 (6):1767–1771. 10.1093/nar/gkp1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Handsaker B, Wysoker A et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 (16):2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuller CW, Middendorf LR, Benner SA et al. (2009) The challenges of sequencing by synthesis. Nat Biotechnol 27(11):1013–1023. 10.1038/nbt.1585 [DOI] [PubMed] [Google Scholar]
- 28.Kim D, Pertea G, Trapnell C et al. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14(4):R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Singh D, Zeng Z et al. (2010) MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 38(18):e178 10.1093/nar/gkq622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A, Davis CA, Schlesinger F et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu TD, Reeder J, Lawrence M et al. (2016) GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. Methods Mol Biol 1418:283–334. 10.1007/978-1-4939-3578-9_15 [DOI] [PubMed] [Google Scholar]
- 32.Lunter G, Goodson M (2011) Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21(6):936–939. 10.1101/gr.111120.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Ruan J, Durbin R (2008) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18(11):1851–1858. 10.1101/gr.078212.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 (14):1754–1760. 10.1093/bioinformatics/btp324. btp324 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, Williams BA, Pertea G et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pertea M, Pertea GM, Antonescu CM et al. (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas BJ, Papanicolaou A, Yassour et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8(8):1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grabherr MG, Haas BJ, Yassour M et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulz MH, Zerbino DR, Vingron M et al. (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28(8):1086–1092. 10.1093/bioinformatics/bts094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patro R, Mount SM, Kingsford C (2014) Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol 32 (5):462–464. 10.1038/nbt.2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30(7):923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 44.Anders S, Pyl PT, Huber W (2015) HTSeq— a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bullard JH, Purdom E, Hansen KD et al. (2010) Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11:94 10.1186/1471-2105-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang L, Schlesinger F, Davis CA et al. (2011) Synthetic spike-in standards for RNA-seq experiments. Genome Res 21 (9):1543–1551. 10.1101/gr.121095.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mortazavi A, Williams BA, McCue K et al. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5 (7):621–628. 10.1038/nmeth.1226. nmeth.1226 [pii] [DOI] [PubMed] [Google Scholar]
- 48.Leek JT, Scharpf RB, Bravo HC et al. (2010) Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev 11(10):733–739. 10.1038/nrg2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leek JT, Storey JD (2007) Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 3 (9):1724–1735. 10.1371/journal.pgen.0030161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Risso D, Ngai J, Speed TP et al. (2014) Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 32(9):896–902. 10.1038/nbt.2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen KD, Wu Z, Irizarry RA et al. (2011) Sequencing technology does not eliminate biological variability. Nat Biotechnol 29 (7):572–573. 10.1038/nbt.1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11(3):R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smyth GK (2005) limma: linear models for microarray data In: Gentleman R, Carey V, Huber, Irizarry R, Dudoit S (eds) Bioinformatics and computational biology solutions using R and Bioconductor. Springer, Berlin, pp 397–420 [Google Scholar]
- 55.Bolstad BM, Irizarry RA, Astrand M et al. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19(2):185–193 [DOI] [PubMed] [Google Scholar]
- 56.Pickrell JK, Marioni JC, Pai AA et al. (2010) Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464(7289):768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li B, Ruotti V, Stewart RM et al. (2010) RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26 (4):493–500. 10.1093/bioinformatics/btp692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner GP, Kin K, Lynch VJ (2012) Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131 (4):281–285. 10.1007/s12064-012-0162-3 [DOI] [PubMed] [Google Scholar]
- 59.Conesa A, Madrigal P, Tarazona S et al. (2016) A survey of best practices for RNA-seq data analysis. Genome Biol 17:13 10.1186/s13059-016-0881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oshlack A, Wakefield MJ (2009) Transcript length bias in RNA-seq data confounds systems biology. Biol Direct 4:14 10.1186/1745-6150-4-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1):118–127. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 62.Karpievitch YV, Nikolic SB, Wilson R et al. (2014) Metabolomics data normalization with EigenMS. PLoS One 9(12):e116221 10.1371/journal.pone.0116221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tracy CA, Widom H (1994) Level spacing distributions and the Bessel kernel. Commun Math Phys 161(2):289–309 [Google Scholar]
- 64.Johnstone IM (2001) On the distribution of the largest eigenvalue in principal components analysis. Ann Stat 29(2):295–327 [Google Scholar]
- 65.Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2(12):e190 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abbas-Aghababazadeh F, Li Q, Fridley BL (2018) Comparison of normalization approaches for gene expression studies completed with high-throughput sequencing. PLoS One 13(10):e0206312 10.1371/journal.pone.0206312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L, Feng Z, Wang X et al. (2010) DEG-seq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26(1):136–138. 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- 68.Langmead B, Hansen KD, Leek JT (2010) Cloud-scale RNA-sequencing differential expression analysis with Myrna. Genome Biol 11(8):R83 10.1186/gb-2010-11-8-r83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J, Witten DM, Johnstone IM et al. (2012) Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics 13(3):523–538. 10.1093/biostatistics/kxr031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Auer PL, Doerge RW (2011) A two-stage Poisson model for testing RNA-seq data. Stat Appl Genet Mol Biol 10(1): [Google Scholar]
- 71.Srivastava S, Chen L (2010) A two-parameter generalized Poisson model to improve the analysis of RNA-seq data. Nucleic Acids Res 38(17):e170 10.1093/nar/gkq670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robinson MD, Smyth GK (2007) Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23 (21):2881–2887. 10.1093/bioinformatics/btm453. btm453 [pii] [DOI] [PubMed] [Google Scholar]
- 73.Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (1):139–140. 10.1093/bioinformatics/btp616. btp616 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Y, Schafer DW, Cumbie JS et al. (2011) The NBP negative binomial model for assessing differential gene expression from RNA-Seq. Stat Appl Genet Mol Biol 10(1):24 [Google Scholar]
- 76.Zhou YH, Xia K, Wright FA (2011) A powerful and flexible approach to the analysis of RNA sequence count data. Bioinformatics 27(19):2672–2678. 10.1093/bioinformatics/btr449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van De Wiel MA, Leday GG, Pardo L et al. (2013) Bayesian analysis of RNA sequencing data by estimating multiple shrinkage priors. Biostatistics 14(1):113–128. 10.1093/biostatistics/kxs031 [DOI] [PubMed] [Google Scholar]
- 78.Hardcastle TJ, Kelly KA (2010) baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11:422 10.1186/1471-2105-11-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 80.Ritchie ME, Phipson B, Wu D et al. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Law CW, Chen Y, Shi W et al. (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15(2):R29 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li J, Tibshirani R (2013) Finding consistent patterns: a nonparametric approach for identifying differential expression in RNA-Seq data. Stat Methods Med Res 22(5):519–536. 10.1177/0962280211428386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tarazona S, Garcia-Alcalde F, Dopazo J et al. (2011) Differential expression in RNA-seq: a matter of depth. Genome Res 21 (12):2213–2223. 10.1101/gr.124321.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodol 57(1):289–300 [Google Scholar]
- 85.Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100(16):9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Storey JD (2002) A direct approach to false discovery rates. J R Stat Soc B Methodol 64 (Pt. 3):479–498 [Google Scholar]
- 87.Bland JM, Altman DG (1995) Multiple significance tests: the Bonferroni method. BMJ 310(6973):170 10.1136/bmj.310.6973.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70 [Google Scholar]
- 89.Hochberg Y (1988) A sharper Bonferroni procedure for multiple tests of significance. Biometrika 75(4):800–802 [Google Scholar]
- 90.Newman AM, Liu CL, Green MR et al. (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12 (5):453–457. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thorsson V, Gibbs DL, Brown SD et al. (2018) The immune landscape of cancer. Immunity 48(4):812–830.e814. 10.1016/j.immuni.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li T, Fan J, Wang B et al. (2017) TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 77(21):e108–e110. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aran D, Hu Z, Butte AJ (2017) xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 18(1):220 10.1186/s13059-017-1349-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hashimshony T, Wagner F, Sher N et al. (2012) CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep 2 (3):666–673. 10.1016/j.celrep.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 95.Islam S, Zeisel A, Joost S et al. (2014) Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods 11(2):163–166. 10.1038/nmeth.2772 [DOI] [PubMed] [Google Scholar]
- 96.Picelli S, Faridani OR, Bjorklund AK et al. (2014) Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9 (1):171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- 97.Macosko EZ, Basu A, Satija R et al. (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161(5):1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zheng GX, Terry JM, Belgrader P et al. (2017) Massively parallel digital transcriptional profiling of single cells. Nat Commun 8:14049 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ziegenhain C, Vieth B, Parekh S et al. (2017) Comparative analysis of single-cell RNA sequencing methods. Mol Cell 65 (4):631–643.e634. 10.1016/j.molcel.2017.01.023 [DOI] [PubMed] [Google Scholar]
- 100.Svensson V, Natarajan KN, Ly LH et al. (2017) Power analysis of single-cell RNA-sequencing experiments. Nat Methods 14 (4):381–387. 10.1038/nmeth.4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ilicic T, Kim JK, Kolodziejczyk AA et al. (2016) Classification of low quality cells from single-cell RNA-seq data. Genome Biol 17:29 10.1186/s13059-016-0888-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lun AT, McCarthy DJ, Marioni JC (2016) A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5:2122 10.12688/f1000research.9501.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Satija R, Farrell JA, Gennert D et al. (2015) Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33 (5):495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao C, Hu S, Huo X et al. (2017) Dr.seq2: a quality control and analysis pipeline for parallel single cell transcriptome and epigenome data. PLoS One 12(7):e0180583 10.1371/journal.pone.0180583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McCarthy DJ, Campbell KR, Lun AT et al. (2017) Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33(8):1179–1186. 10.1093/bioinformatics/btw777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Finak G, McDavid A, Yajima M et al. (2015) MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16:278 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lun AT, Bach K, Marioni JC (2016) Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol 17:75 10.1186/s13059-016-0947-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kharchenko PV, Silberstein L, Scadden DT (2014) Bayesian approach to single-cell differential expression analysis. Nat Methods 11 (7):740–742. 10.1038/nmeth.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang Y, Zhang NR, Li M (2017) SCALE: modeling allele-specific gene expression by single-cell RNA sequencing. Genome Biol 18(1):74 10.1186/s13059-017-1200-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu Z, Lou H, Xie K et al. (2017) Reconstructing cell cycle pseudo time-series via single-cell transcriptome data. Nat Commun 8(1):22 10.1038/s41467-017-00039-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McDavid A, Finak G, Gottardo R (2016) The contribution of cell cycle to heterogeneity in single-cell RNA-seq data. Nat Biotechnol 34 (6):591–593. 10.1038/nbt.3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang J, Huang M, Torre E et al. (2018) Gene expression distribution deconvolution in single-cell RNA sequencing. Proc Natl Acad Sci U S A 115(28):E6437–E6446. 10.1073/pnas.1721085115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vallejos CA, Risso D, Scialdone A et al. (2017) Normalizing single-cell RNA sequencing data: challenges and opportunities. Nat Methods 14(6):565–571. 10.1038/nmeth.4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cole MB, Risso D, Wagner A et al. (2019) Performance assessment and selection of normalization procedures for single-cell RNA--Seq. Cell Syst 8(4):315–328.e318. 10.1016/j.cels.2019.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bacher R, Chu LF, Leng N et al. (2017) SCnorm: robust normalization of single-cell RNA-seq data. Nat Methods 14(6):584–586. 10.1038/nmeth.4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jia C, Hu Y, Kelly D et al. (2017) Accounting for technical noise in differential expression analysis of single-cell RNA sequencing data. Nucleic Acids Res 45(19):10978–10988. 10.1093/nar/gkx754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vallejos CA, Marioni JC, Richardson S (2015) BASiCS: Bayesian analysis of single-cell sequencing data. PLoS Comput Biol 11 (6):e1004333 10.1371/journal.pcbi.1004333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Prabhakaran S, Azizi E, Carr A et al. (2016) Dirichlet process mixture model for correcting technical variation in single-cell gene expression data. JMLR Workshop Conf Proc 48:1070–1079 [PMC free article] [PubMed] [Google Scholar]
- 119.Azizi E, Prabhakaran S, Carr A et al. (2017) Bayesian inference for single-cell clustering and imputing. Genomics Comput Biol 3(1): e46 10.18547/gcb.2017.vol3.iss1.e46 [DOI] [Google Scholar]
- 120.Gong W, Kwak IY, Pota P et al. (2018) DrImpute: imputing dropout events in single cell RNA sequencing data. BMC Bioinformatics 19(1):220 10.1186/s12859-018-2226-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang M, Wang J, Torre E et al. (2018) SAVER: gene expression recovery for single-cell RNA sequencing. Nat Methods 15 (7):539–542. 10.1038/s41592-018-0033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mongia A, Sengupta D, Majumdar A (2019) McImpute: matrix completion based imputation for single cell RNA-seq data. Front Genet 10:9 10.3389/fgene.2019.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li WV, Li JJ (2018) An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat Commun 9(1):997 10.1038/s41467-018-03405-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Linderman GC, Zhao J, Kluger Y (2018) Zero-preserving imputation of scRNA-seq data using low-rank approximation. bioRxiv:397588 10.1101/397588 [DOI] [Google Scholar]
- 125.Chen C, Wu C, Wu L et al. (2018) scRMD: imputation for single cell RNA-seq data via robust matrix decomposition. bioRxiv:459404 10.1101/459404 [DOI] [PubMed] [Google Scholar]
- 126.van Dijk D, Sharma R, Nainys J et al. (2018) Recovering gene interactions from single-cell data using data diffusion. Cell 174 (3):716–729.e727. 10.1016/j.cell.2018.05.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ronen J, Akalin A (2018) netSmooth: network-smoothing based imputation for single cell RNA-seq. F1000Res 7:8 10.12688/f1000research.13511.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wagner F, Yan Y, Yanai I (2017) K-nearest neighbor smoothing for high-throughput single-cell RNA-Seq data. bioRxiv:217737 10.1101/217737 [DOI] [Google Scholar]
- 129.Zhang L, Zhang S (2018) Comparison of computational methods for imputing single-cell RNA-sequencing data. IEEE/ACM Trans Comput Biol Bioinform. 10.1109/TCBB.2018.2848633 [DOI] [PubMed] [Google Scholar]
- 130.Andrews TS, Hemberg M (2018) False signals induced by single-cell imputation. F1000Res 7:1740 10.12688/f1000research.16613.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Buettner F, Natarajan KN, Casale FP et al. (2015) Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol 33(2):155–160. 10.1038/nbt.3102 [DOI] [PubMed] [Google Scholar]
- 132.Katayama S, Tohonen V, Linnarsson S et al. (2013) SAMstrt: statistical test for differential expression in single-cell transcriptome with spike-in normalization. Bioinformatics 29 (22):2943–2945. 10.1093/bioinformatics/btt511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ding B, Zheng L, Zhu Y et al. (2015) Normalization and noise reduction for single cell RNA-seq experiments. Bioinformatics 31 (13):2225–2227. 10.1093/bioinformatics/btv122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lun ATL, Calero-Nieto FJ, Haim-Vilmovsky L et al. (2017) Assessing the reliability of spike-in normalization for analyses of single-cell RNA sequencing data. Genome Res 27 (11):1795–1806. 10.1101/gr.222877.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vieth B, Parekh S, Ziegenhain C et al. (2019) A systematic evaluation of single cell RNA-Seq analysis pipelines: library preparation and normalisation methods have the biggest impact on the performance of scRNA-seq studies. bioRxiv:583013 10.1101/583013 [DOI] [Google Scholar]
- 136.Buttner M, Miao Z, Wolf FA et al. (2019) A test metric for assessing single-cell RNA-seq batch correction. Nat Methods 16(1):43–49. 10.1038/s41592-018-0254-1 [DOI] [PubMed] [Google Scholar]
- 137.Haghverdi L, Lun ATL, Morgan MD et al. (2018) Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36(5):421–427. 10.1038/nbt.4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stuart T, Butler A, Hoffman P et al. (2018) Comprehensive integration of single cell data. bioRxiv:460147 10.1101/460147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kiselev VY, Andrews TS, Hemberg M (2019) Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev 20 (5):273–282. 10.1038/s41576-018-0088-9 [DOI] [PubMed] [Google Scholar]
- 140.Brennecke P, Anders S, Kim JK et al. (2013) Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods 10 (11):1093–1095. 10.1038/nmeth.2645 [DOI] [PubMed] [Google Scholar]
- 141.Fan J, Salathia N, Liu R et al. (2016) Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat Methods 13(3):241–244. 10.1038/nmeth.3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Usoskin D, Furlan A, Islam S et al. (2015) Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18(1):145–153. 10.1038/nn.3881 [DOI] [PubMed] [Google Scholar]
- 143.Hyvarinen A, Oja E (2000) Independent component analysis: algorithms and applications. Neural Netw 13(4–5):411–430 [DOI] [PubMed] [Google Scholar]
- 144.Belkin M, Niyogi P (2003) Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput 15 (6):1373–1396. 10.1162/089976603321780317 [DOI] [Google Scholar]
- 145.Van Der Maaten L, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9:2579–2605 [Google Scholar]
- 146.Hicks SC, Townes FW, Teng M et al. (2018) Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics 19(4):562–578. 10.1093/biostatistics/kxx053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Risso D, Perraudeau F, Gribkova S et al. (2018) A general and flexible method for signal extraction from single-cell RNA-seq data. Nat Commun 9(1):284 10.1038/s41467-017-02554-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kobak D, Berens P (2018) The art of using t-SNE for single-cell transcriptomics. bioRxiv:453449 10.1101/453449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wattenberg M, Viegas F, Johnson I (2016) How to use t-SNE effectively. Distill.pub 10.23915/distill.00002 [DOI] [Google Scholar]
- 150.Linderman GC, Rachh M, Hoskins JG et al. (2019) Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat Methods 16(3):243–245. 10.1038/s41592-018-0308-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McInnes L, Healy J, Melville J (2018) UMAP: uniform manifold approximation and projection for dimension reduction. arXiv e-prints [Google Scholar]
- 152.Becht E, McInnes L, Healy J et al. (2018) Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37:38 10.1038/nbt.4314. https://www.nature.com/articles/nbt.4314#supplementary-information [DOI] [PubMed] [Google Scholar]
- 153.Tung PY, Blischak JD, Hsiao CJ et al. (2017) Batch effects and the effective design of single-cell gene expression studies. Sci Rep 7:39921 10.1038/srep39921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Andrews TS, Hemberg M (2018) Identifying cell populations with scRNASeq. Mol Asp Med 59:114–122. 10.1016/j.mam.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 155.Navin NE (2014) Cancer genomics: one cell at a time. Genome Biol 15(8):452 10.1186/s13059-014-0452-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang Y, Navin NE (2015) Advances and applications of single-cell sequencing technologies. Mol Cell 58(4):598–609. 10.1016/j.molcel.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Duo A, Robinson MD, Soneson C (2018) A systematic performance evaluation of clustering methods for single-cell RNA-seq data. F1000Res 7:1141 10.12688/f1000research.15666.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kiselev VY, Kirschner K, Schaub MT et al. (2017) SC3: consensus clustering of single-cell RNA-seq data. Nat Methods 14 (5):483–486. 10.1038/nmeth.4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang B, Zhu J, Pierson E et al. (2017) Visualization and analysis of single-cell RNA-seq data by kernel-based similarity learning. Nat Methods 14(4):414–416. 10.1038/nmeth.4207 [DOI] [PubMed] [Google Scholar]
- 160.Grun D, Lyubimova A, Kester L et al. (2015) Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525 (7568):251–255. 10.1038/nature14966 [DOI] [PubMed] [Google Scholar]
- 161.Zurauskiene J, Yau C (2016) pcaReduce: hierarchical clustering of single cell transcriptional profiles. BMC Bioinformatics 17:140 10.1186/s12859-016-0984-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lin P, Troup M, Ho JW (2017) CIDR: ultrafast and accurate clustering through imputation for single-cell RNA-seq data. Genome Biol 18(1):59 10.1186/s13059-017-1188-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zeisel A, Munoz-Manchado AB, Codeluppi S et al. (2015) Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347 (6226):1138–1142. 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]
- 164.Guo M, Wang H, Potter SS et al. (2015) SINCERA: a pipeline for single-cell RNA-Seq profiling analysis. PLoS Comput Biol 11(11): e1004575 10.1371/journal.pcbi.1004575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen J, Schlitzer A, Chakarov S et al. (2016) Mpath maps multi-branching single-cell trajectories revealing progenitor cell progression during development. Nat Commun 7:11988 10.1038/ncomms11988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Senabouth A, Lukowski SW, Alquicira Hernandez J et al. (2017) ascend: R package for analysis of single cell RNA-seq data. bioRxiv:207704 10.1101/207704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ester M, Kriegel H-P, et al. (1996) A density-based algorithm for discovering clusters a density-based algorithm for discovering clusters in large spatial databases with noise. Paper presented at the Proceedings of the Second International Conference on Knowledge discovery and data mining, Portland, Oregon [Google Scholar]
- 168.Jiang L, Chen H, Pinello L et al. (2016) GiniClust: detecting rare cell types from single-cell gene expression data with Gini index. Genome Biol 17(1):144 10.1186/s13059-016-1010-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Trapnell C, Cacchiarelli D, Grimsby J et al. (2014) The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32 (4):381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rosvall M, Bergstrom CT (2008) Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A 105(4):1118–1123. 10.1073/pnas.0706851105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Blondel VD, Guillaume J-L, Lambiotte R et al. (2008) Fast unfolding of communities in large networks. J Stat Mech Theory Exp 2008:10008 [Google Scholar]
- 172.Lancichinetti A, Fortunato S (2009) Community detection algorithms: a comparative analysis. Phys Rev E 80(5):056117 10.1103/PhysRevE.80.056117 [DOI] [PubMed] [Google Scholar]
- 173.Levine JH, Simonds EF, Bendall SC et al. (2015) Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162(1):184–197. 10.1016/j.cell.2015.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ding J, Shah S, Condon A (2016) densityCut: an efficient and versatile topological approach for automatic clustering of biological data. Bioinformatics 32 (17):2567–2576. 10.1093/bioinformatics/btw227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xu C, Su Z (2015) Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics 31 (12):1974–1980. 10.1093/bioinformatics/btv088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wolf FA, Angerer P, Theis FJ (2018) SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19(1):15 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Baran Y, Sebe-Pedros A, Lubling Y et al. (2018) MetaCell: analysis of single cell RNA-seq data using k-NN graph partitions. bioRxiv:437665 10.1101/437665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Xie P, Gao M, Wang C et al. (2019) SuperCT: a supervised-learning framework for enhanced characterization of single-cell transcriptomic profiles. Nucleic Acids Res. 10.1093/nar/gkz116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Aran D, Looney AP, Liu L et al. (2019) Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20(2):163–172. 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li J, Smalley I, Schell MJ et al. (2017) SinCHet: a MATLAB toolbox for single cell heterogeneity analysis in cancer. Bioinformatics 33(18):2951–2953. 10.1093/bioinformatics/btx297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ferrall-Fairbanks MC, Ball M, Padron E et al. (2019) Leveraging single-cell RNA sequencing experiments to model intratumor heterogeneity. JCO Clin Cancer Informatics 3:1–10. 10.1200/cci.18.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yang X, Liu D, Liu F et al. (2013) HTQC: a fast quality control toolkit for Illumina sequencing data. BMC Bioinformatics 14:33 10.1186/1471-2105-14-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7(2):e30619 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 (15):2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cox MP, Peterson DA, Biggs PJ (2010) SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485 10.1186/1471-2105-11-485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Robertson G, Schein J, Chiu R et al. (2010) De novo assembly and analysis of RNA-seq data. Nat Methods 7(11):909–912. 10.1038/nmeth.1517 [DOI] [PubMed] [Google Scholar]