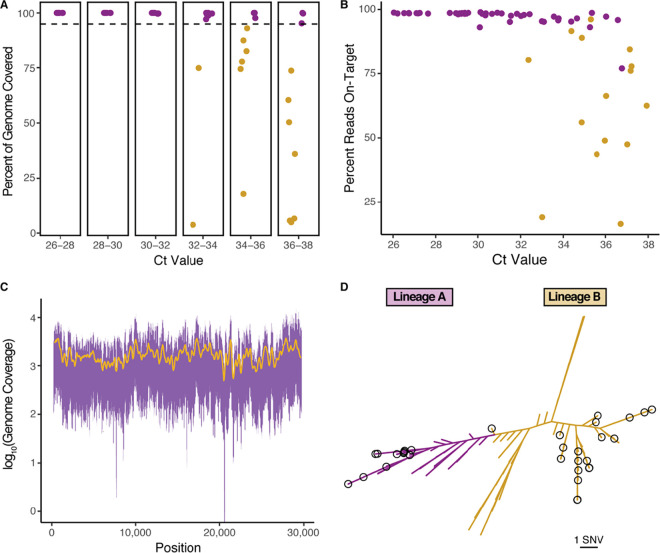
FIG 1.
Evaluation of the Swift Biosciences’ SARS-CoV-2 multiplex amplicon sequencing panel. (A) Complete genomes were recovered from all samples with a CT value of ≤32.16 and a CT value as high as 36.77. Samples for which complete genomes (>95% genome coverage) were recovered are highlighted in purple. Partial genomes are highlighted in gold. (B) SARS-CoV-2 sequences were highly enriched in the sequencing libraries as measured by the percentage of reads mapping to the reference genome for SARS-CoV-2 (NCBI accession no. NC_045512.2). Complete genomes were recovered for samples highlighted in purple, while partial genomes were recovered for those highlighted in gold. (C) The genome coverage between nucleotides 201 and 29741 of the SARS-CoV-2 reference genome is even. The 5th and 95th percentiles of coverage at each position across the 41 samples with a mean depth of >100× are plotted in purple. A 250-nucleotide window moving average is represented in gold. (D) The 46 SARS-CoV-2 samples with complete genomes belong to both major SARS-CoV-2 lineages. A phylogenetic tree with the 46 SARS-CoV-2 genomes recovered in this report and 109 other global strains was constructed with FastTree version 2.1.1. Strains belonging to lineage A are highlighted in purple, while those belonging to lineage B are highlighted in gold. Those genomes sequenced in this report are circled in black. SNV, single nucleotide variation.