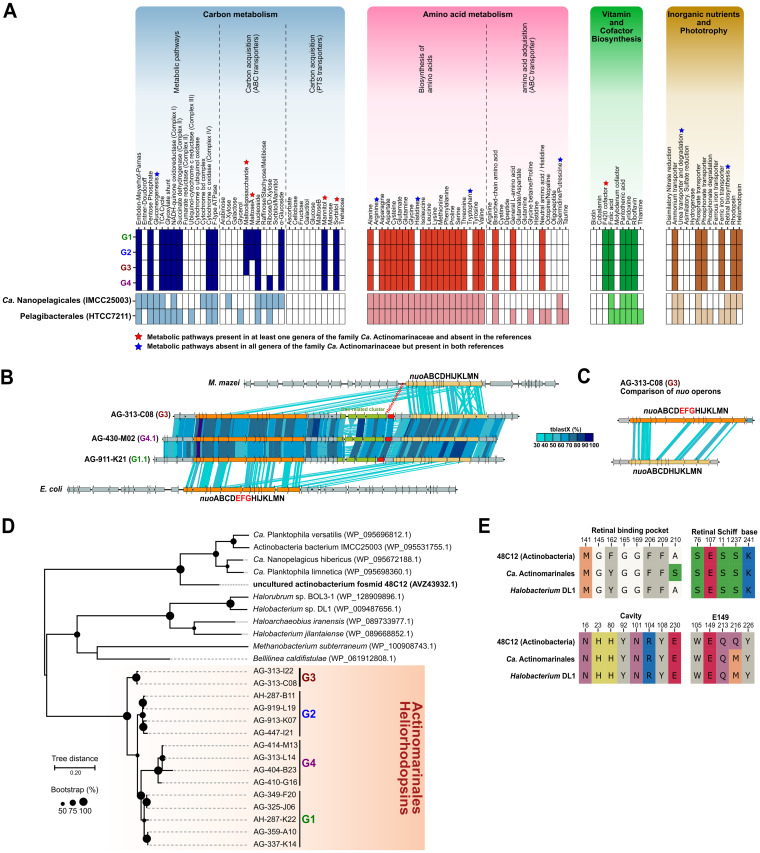
FIG 5.
(A) Inferred metabolism of the four “Ca. Actinomarinales” genera based on the KEGG database. “Ca. Nanopelagicales” (IMCC25003) and Pelagibacterales (HTCC7211) were added for the comparison against other streamlined genomes. (B) Genomic alignment (in amino acids) of the two nuo paralogs close to the heliorhodopsin. nuo operons of Escherichia coli and Methanosarcina mazei were added for the comparison. (C) Alignment (in amino acids) between the two nuo paralogs detected in “Ca. Actinomarinales”, indicating that the subunits EFG were missing in one copy. (D) Maximum likelihood phylogenetic tree of the heliorhodopsin protein. Accession numbers for the reference heliorhodopsins are indicated within parentheses. Bootstrap values are indicated as black circles on the nodes. (E) Conserved key residues among the reference 48C12, retrieved from a fosmid of an uncultured freshwater Actinobacteria (GenBank accession number MF737519), Halobacterium DL1, and “Ca. Actinomarinales” heliorhodopsins.