Abstract
COVID-19 has caused extensive human casualties with significant economic impacts around the globe, and has imposed new challenges on health systems worldwide. Over the past decade, SARS, Ebola, and Zika also led to significant concerns among the scientific community. Interestingly, the SARS and Zika epidemics ended before vaccine development; however, the scholarly community and the pharmaceutical companies responded very quickly at that time. Similarly, when the genetic sequence of SARS-CoV-2 was revealed, global vaccine companies and scientists have stepped forward to develop a vaccine, triggering a race toward vaccine development that the whole world is relying on. Similarly, an effective and safe vaccine could play a pivotal role in eradicating COVID-19. However, few important questions regarding SARS-CoV-2 vaccine development are explored in this review.
Keywords: COVID-19, Vaccine, Vaccine backfires, Vaccine safety
INTRODUCTION
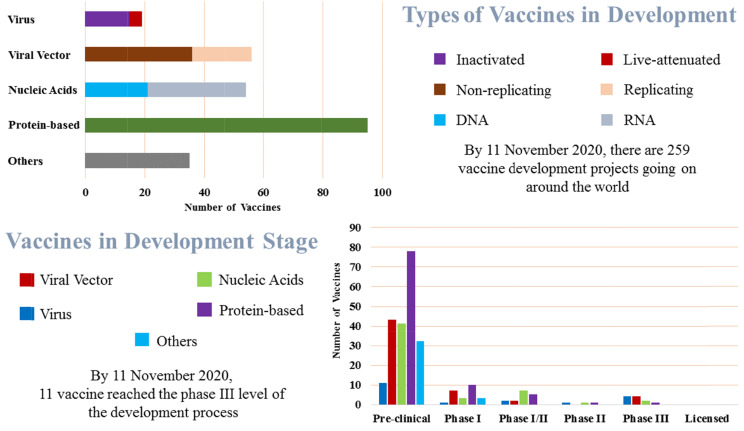
Humans have a long history of battling against the viruses. The constant battle between viruses and scientists has been recognized as a key driver of medical advances. In this series of battles, the latest one is the outbreak of a novel coronavirus, COVID-19, originating in the Wuhan region of China in late December 2019 (Ratan et al., 2020). COVID-19 has affected at least 216 countries, areas or territories around the world. So far, there had been approximately 54 million 301 thousand 156 cases and one million 316 thousand 994 deaths confirmed globally due to this deadly virus by November 16, 2020 (World Health Organization, 2020). Prior to the emergence of Severe Acute Respiratory Syndrome CoV-2 (SARS-CoV-2), there were six human CoVs (HCoVs), including that which caused the SARS global outbreak that started in November 2002 in the Guangdong province of China. After that epidemic, China reported more than 8,000 cases of disease and 774 deaths with a case-fatality rate of 7%. A decade later in 2012, Middle East respiratory syndrome CoV (MERS-CoV) first emerged in Saudi Arabia, with a total of 2,494 laboratory-confirmed cases and 858 deaths with a case-fatality ratio of 34.4% (Corman et al., 2018; Peeri et al., 2020). However, COVID-19 has caused a pandemic that has compelled the global economy to grind to a halt. A vaccine remains the best option for restoring normal life and global economies. This has triggered a vaccine development race. According to Mullard’s report (Mullard, 2020), as of 11 November 2020, there were 259 COVID-19 vaccine projects going on around the world. Of those, there were 79 protein, 16 virus-like particle, 22 DNA, 33 RNA, 36 non-replicating and 20 replicating viral vector, 15 inactivated and four live-attenuated, and 35 other vaccines candidate in development pipeline. Among them, 204 were still in the preclinical stage; 11 candidates were reached phase III clinical trial stage, and the remaining were in between these stages (Fig. 1) (London School of Hygiene and Tropical Medicine, 2020).
Fig. 1.
State of the vaccine race for COVID-19.
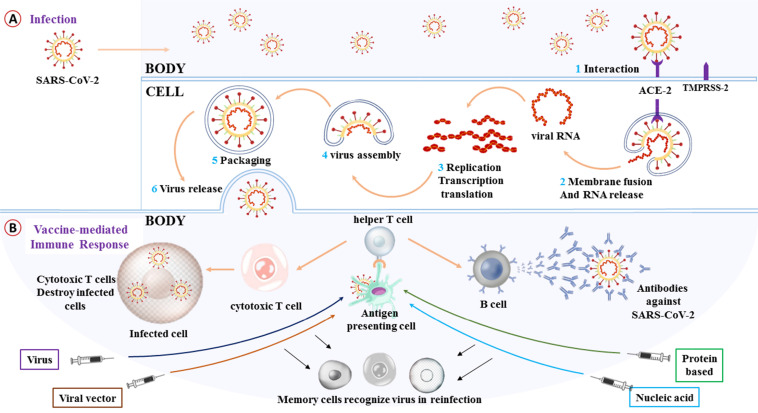
A vaccine introduces the structure and biological agents of a specific virus to antigen-presenting cells of the host, which engulf it and pass portions of it to activate helper T (Th) cells. The Th cells then trigger other immune responses i.e., activation of B cells and cytotoxic T (Tc) cells. B cells produce antibodies that can prevent the virus from infecting cells, while Tc cells recognize and kill cells that are infected with the virus, that help the surveillance cells of the body to track the virus for long periods (Fig. 2B). In principle, understanding the etiology, epidemiology, pathogenesis and immunobiology of the infection is of the utmost importance for the development of vaccines (Zepp, 2010). Thus, a few simple questions, although complicated to answer, have arisen regarding the basic principles of vaccines that need to be resolved with regard to COVID-19 vaccine development. Here, we aim to address those simple questions.
Fig. 2.
COVID-19 and the vaccine. (A) A simple representation of the COVID-19 infection mechanism in the body. (B) Basic principles of vaccines in generalized form.
STRUCTURE AND PATHOPHYSIOLOGY OF COVID-19
SARS-CoV-2 is a β-coronavirus belonging to the Sarbecovirus subgenus of the Coronaviridae family, and is enveloped with non-segmented positive-sense RNA virus (Zhu et al., 2020). In broad terms, the genome of this virus can be divided into two parts. The first open reading frame (ORF 1a/b) comprises two-thirds of the total viral genome (~30 kb) and encodes 16 non-structure proteins. This ORF 1a/b has the genetic function to roll-out the viral replication that controls the production of cellular proteins and keeps evading the immune system of the host. The remaining portion of the genome codes for four basic structural proteins, including spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein, and several other accessory proteins (Guo et al., 2020). The crucial part of SARS-CoV-19 and COVID-19 infection begins when the S protein of this pathogen binds to angiotensin-converting enzyme 2 (ACE2), a cellular receptor of the host (Fig. 2A). After binding to the ACE2 receptor, the conformation shift in the S protein enables fusion of the viral envelope with the cell membrane via the endosomal pathway. Then, the virus enters the cell and releases its RNA. This RNA then goes through transcription, translation, and replication. In this production line, RNA is translated into replicase polyproteins pp1a and 1ab, and those are then severed by viral proteinase into small products. Polymerase by discontinuous transcription yields a sequence of subgenomic mRNAs that are eventually transformed into specific viral proteins. Subsequently, in the endoplasmic reticulum and Golgi apparatus, viral proteins and genome RNA are packaged into the virion and then transported through vesicles to be released from the cell (Shereen et al., 2020).
TYPES OF VACCINE
The Coalition for Epidemic Preparedness Innovations (CEPI), a multilateral and multinational stakeholders foundation for the development of vaccine against infectious diseases, informed in September 2020 that the nine separate technological platforms are being used to make an effective vaccine against SARS-CoV-2 (Gopinathan et al., 2020). Scientist around the world using both the classical and next-generation platforms. The classical platforms are whole-inactivated virus, live-attenuated virus, protein subunit, and virus-like particles, and the next-generation platforms are nucleic acids (RNA and DNA), viral vectors (non-replicating and replicating), recombinant protein, and antigen-presenting cells (Le et al., 2020; van Riel and de Wit, 2020). However, as of September 2020 reported by CEPI, most of the frontrunner vaccines in clinical studies have emphasized on spike protein of coronavirus and its versions as the key antigen responsible for the infection of COVID-19. CEPI also pointed out that eleven candidate vaccine in clinical stage using adjuvant to improve the immunogenicity (Le et al., 2020).
Among the ongoing or planned clinical trial as of 11 November 2020, double-blind, single-blind, dose-confirmation, observer-blind randomized, and open-label non-randomized studies are designed for different types of the vaccine from the various technological platform, and of them, there are 18 nucleic acids, 8 Non-replicating viral vectors, 2 replicating viral vector, 9 inactivated virus, 1 virus-like particle, 11 protein subunit, and 3 other candidate vaccines. Moreover, on this clinical trial tally, China is on the top followed by the USA, Australia, Canada, UK, and others (Table 1).
Table 1.
Ongoing or planned clinical trial for COVID-19 vaccine up to 11 November 2020
| Candidate | Type | Phase | Study design | Volunteer | Country | Reference |
|---|---|---|---|---|---|---|
| Moderna mRNA-1273 | RNA | 3 | Double-blind randomized | 30000 | USA | NCT04470427 |
| WIBP vaccine | Inactivated | 3 | Double-blind randomized | 45,000 | Bahrain, Jordan, Egypt, UAE | NCT04510207 |
| Sinovac CoronaVac | Inactivated | 3 | Double-blind randomized | 13,060 | Brazil | NCT04456595 |
| Oxford ChAdOx1-S | Non-replicating viral vector | 3 | Double-blind randomized | 40,051 | USA, Chile, Peru | NCT04516746 |
| Novavax NVX-CoV2373 | Protein subunit | 3 | Double-blind randomized | 30,000 | USA, Mexico, Puerto Rico | NCT04611802 |
| Novavax NVX-CoV2373 | Protein subunit | 3 | Double-blind randomized | 9,000 | UK | NCT04583995 |
| Moderna mRNA-1273 | RNA | 3 | Double-blind randomized | 30,000 | USA | NCT04470427 |
| Cansino Ad5-nCoV | Non-replicating viral vector | 3 | Double-blind randomized | 40,000 | Pakistan | NCT04526990 |
| BIBP/ Sinopharm BBIBP-CorV | Inactivated | 3 | Double-blind randomized | 3,000 | Argentina | NCT04560881 |
| Oxford ChAdOx1-S | Non-replicating viral vector | 2/3 | Single-blind randomized | 12,390 | UK | NCT04400838 |
| BioNTech BNT162 | RNA | 2/3 | Dose-finding, double-blind randomized | 43,998 | USA, Argentina, Brazil, others | NCT04368728 |
| BioNTech BNT162 | RNA | 2/3 | Dose-finding, open-label non-randomized | 43,998 | USA, Argentina, Brazil, others | NCT04380701 |
| AZLB protein subunit vaccine | Protein subunit | 2 | Double-blind randomized | 900 | China | NCT04466085 |
| Novavax NVX-CoV2373 | Protein subunit | 2 | Single-blind randomized | 4,400 | South Africa | NCT04533399 |
| Curevac CVnCoV | RNA | 2 | Dose-confirmation,double-blind, randomized | 691 | Peru | NCT04515147 |
| Oxford ChAdOx1-S | Non-replicating viral vector | 1/2 | Double-blind randomized | 2000 | South Africa | NCT04444674 |
| WIBP vaccine | Inactivated | 1/2 | Dose-finding, double-blind randomized | 1,264 | China | ChiCTR2000031809 |
| Bharat Covaxin | Inactivated | 1/2 | Double-blind randomized | 755 | India | NCT04471519 |
| Oxford ChAdOx1-S | Non-replicating viral vector | 1/2 | Single-blind randomized | 1,090 | UK | NCT04324606 |
| Zydus Cadila ZyCoV-D | DNA | 1/2 | Double-blind randomized | 1048 | India | CTRI/2020/07/026352 |
| CAMS vaccine | Inactivated | 1/2 | Dose-finding, double-blind randomized | 942 | China | NCT04412538 |
| Sinovac CoronaVac | Inactivated | 1/2 | Dose-finding, double-blind randomized | 744 | China | NCT04352608 |
| Cansino Ad5-nCoV | Non-replicating viral vector | 1/2 | Dose-finding, double-blind randomized | 696 | Canada | NCT04398147 |
| CAMS vaccine | Inactivated | 1/2 | Double-blind randomized | 471 | China | NCT04470609 |
| Sinovac CoronaVac | Inactivated | 1/2 | Dose-finding, double-blind randomized | 422 | China | NCT04383574 |
| Genexine GX-19 | DNA | 1/2 | Dose-finding, double-blind randomized | 210 | Republic of Korea | NCT04445389 |
| Aivita AV-COVID-19 | Other | 1/2 | Dose-finding, double-blind randomized | 180 | USA | NCT04386252 |
| KBP-COVID-19 | Protein subunit | 1/2 | Observer-blind, dose-finding randomized | 180 | Not Provided | NCT04473690 |
| Inovio INO-4800 | DNA | 1/2 | Dose-finding, Open-label (A), double-blind (B) randomized | 160 | Republic of Korea | NCT04447781 |
| Arcturus ARCT-021 | RNA | 1/2 | Double-blind randomized | 92 | Singapore | NCT04480957 |
| AnGes AG0301-COVID19 | DNA | 1/2 | Dose-finding, Open-label non-randomized | 30 | Japan | NCT04463472 |
| Themis V591 | Replicating viral vector | 1/2 | Dose-finding, double-blind randomized | 260 | USA, Austria, Belgium | NCT04498247 |
| Inovio INO-4800 | DNA | 1/2 | Dose-finding, open-label (A), double-blind (B) randomized | 160 | Republic of Korea | NCT04447781 |
| Novavax NVX-CoV2373 | Protein subunit | 1/2 | Dose-finding, observer-blind randomized | 1,419 | Australia, USA | NCT04368988 |
| Imperial LNP-nCoVsaRNA | RNA | 1 | Dose-finding partially randomized | 320 | UK | ISRCTN17072692 |
| Medicago CoVLP | Virus-like particle | 1 | Dose-finding, open-label randomized | 180 | Canada | NCT04450004 |
| Curevac CVnCoV | RNA | 1 | Dose-finding, single-blind randomized | 284 | Belgium, Germany | NCT04449276 |
| PLA-AMS ARCoV | RNA | 1 | Dose-finding randomized | 168 | China | ChiCTR2000034112 |
| Moderna mRNA-1273 | RNA | 1 | Dose-finding, open-label non-randomized | 120 | USA | NCT04283461 |
| Clover SCB-2019 | Protein subunit | 1 | Dose-finding, double-blind randomized | 150 | Australia | NCT04405908 |
| BioNTech BNT162 | RNA | 1 | Double-blind randomized | 144 | China | NCT04523571 |
| Inovio INO-4800 | DNA | 1 | Dose-finding, open-label non-randomized | 120 | USA | NCT04336410 |
| University of Queensland vaccine | Protein subunit | 1 | Dose-finding, double-blind randomized | 216 | Australia | NCT04495933 |
| Symvivo bacTRL-Spike | DNA | 1 | Dose-finding, observer-blind randomized | 12 | Australia | NCT04334980 |
| Cansino Ad5-nCoV | Non-replicating viral vector | 1 | Dose-finding, open-label non-randomized | 108 | China | NCT04313127 |
| SGMI aAPC | Other | 1 | Open-label non-randomized | 100 | China | NCT04299724 |
| SGMI LV-SMENP-DC | Other | 1 | Open-label non-randomized | 100 | China | NCT04276896 |
| Themis V591 | Replicating viral vector | 1 | Dose-finding, double-blind randomized | 90 | Belgium, France | NCT04497298 |
| Gamaleya Gam-COVID-Vac (Lyo) | Non-replicating viral vector | 1 | Open-label non-randomized | 38 | Russia | NCT04437875 |
| AZLB protein subunit vaccine | Protein subunit | 1 | Double-blind randomized | 50 | China | NCT04445194 |
| Medigen MVC-COV1901 | Protein subunit | 1 | Dose finding, open-label non-randomized | 45 | Taiwan | NCT04487210 |
| Vaxine protein subunit vaccine | Protein subunit | 1 | Double-blind randomized | 40 | Australia | NCT04453852 |
aAPC, artificial antigen presenting cell; AZLB, Anhui Zhifei Longcom Biopharmaceutical; BIBP, Beijing Institute of Biological Products; CAMS, Chinese Academy of Medical Sciences; KBP, Kentucky BioProcessing; LV-SMENP-DC, vaccine comprising dendritic cells (DCs) modified with lentivirus (LV) vectors expressing ‘SMENP’ minigene; PLA-AMS, People’s Liberation Army Academy of Military Science; SGMI, Shenzhen Geno-Immune Medical Institute; WIBP, Wuhan Institute of Biological Products.
THE CRITICAL QUESTIONS
The challenge for modern vaccinology is to be able to provoke all the requisite steps leading to immune system activation in vivo, and to provide a non-virulent, harmless type of a given agent capable of generating a strong and adequate immune response tailored against specific viral attack (Moser and Leo, 2010). Thus, some questions arise regarding the development of the vaccine (see Table 1 for current development state of COVID-19 vaccines) that will be administered to billions of people at risk of COVID-19 infection.
WILL VACCINE STIMULATE THE IMMUNE RESPONSE?
As mentioned earlier, ACE2 is the route of SARS-CoV-2 infection. However, this receptor plays a vital role in both innate and adaptive immune responses by modulating the antigen present antigen cells that interact with T cells to initiate defense initiatives (Bernstein et al., 2018). This receptor of transmembrane protease acts in the conversion of angiotensin 1-8 (Ang II) to angiotensin 1-7 (Ang 1-7), prompting diuresis/natriuresis, preserving renal function, and attenuating cardiac and vascular reformation (Vickers et al., 2002; Santos et al., 2008; Zhang et al., 2010). ACE2 also has an important role in the nervous system, and disruption of this receptor can trigger neurological disorders (Kabbani and Olds, 2020). However, a study reported that innate T cells, a heterogeneous class of T lymphocytes (MAIT, γδT and iNKT cells), are also altered by SARS-CoV-2 (Jouan et al., 2020). Besides, a study cohort of 38 patients found that a decline in T cells, B cells, and NK cells was linked to SARS due to coronavirus (Cui et al., 2003). On the other hand, a study conducted on bronchoalveolar lavage fluid of eight COVID-19 patients exhibited chemokine-dominant hypercytokinemia, often called a ‘cytokine storm,’ which robustly promotes expression of numerous IFN-stimulated genes that lead to multi-organ failure (Zhou et al., 2020). Therefore, SARS-CoV-2, directly and indirectly, triggers the impairment and hyper-stimulation of the immune system (Jamilloux et al., 2020; Yazdanpanah et al., 2020). But, cellular immunogenicity, humoral, and cell-mediated immune responses are crucial for vaccine-derived immunity and rapid cytotoxic response against viral infection (Morris et al., 2016; Ewer et al., 2017). Thus, should vaccines stimulate or suppress the immune response system of the host against COVID-19 infections?
WILL A VACCINE PROVIDE SUSTAINABLE IMMUNE ENDURANCE?
Another major concern about immunity against coronaviruses is the endurance of the immune response system. For effective immunization, vaccine-induced long-term regulation of the immune system, especially humoral and cell-mediated arms of the adaptive system, functions through producing the effector cells for the current infection and memory cells for future infections with the pathogenic agent (Clem, 2011). However, a number of studies showed that immune responses against COVID-19 do not last long-term. A study conducted on 285 SARS-CoV-2-infected persons reported that antiviral immunoglobulin-G (IgG) and IgM were increased during the first 3 weeks after symptom onset, and then began to decrease (Long et al., 2020a). A case report of 34 hospitalized patients (admitted from Feb 1 to Feb 29, 2020) with confirmed SARS-CoV-2 revealed that IgM levels reached their peak of after three weeks and then continued to decline up to the end of 7 weeks of observation, whereas IgG values remained more or less the same (Xiao et al., 2020). In another case report, both IgM and the IgG declined after the peak period; this study was conducted on 60 convalescent patients where the value of those two antibodies reached their summit 6-7 weeks after onset, and a decline was observed in the following week (Du et al., 2020). A report from the National COVID Scientific Advisory Panel of the UK mentioned that IgG titers increased within three weeks of the onset of symptoms and started to drop by eight weeks in plasma samples collected from 40 confirmed COVID-infected persons (Adams et al., 2020). Another study noted that the most plasma samples obtained from eight convalescent COVID-19 patients recovering from COVID-19 without hospitalization did not contain high neutralizing activity levels (Robbiani et al., 2020). In cases of asymptomatic infection, a study conducted on 37 individuals reported that they had a weaker immune response, i.e., a greater reduction of IgG and neutralizing antibody levels (Long et al., 2020b). These phenomena are not new to the scientific world. Exactly 30 years ago, in 1990, a study reported a similar result. From an investigation of circulating lymphocyte populations in 15 volunteers infected with a CoV 229-E strain, the researchers observed that the concentration of antibodies began to rise one week after inoculation and then reached their peak another week later. After that, titers of the antibody began to decline. They also claimed that despite the slightly high concentration after one year, this did not always prevent the volunteer from being reinfected with homologous virus (Callow et al., 1990). Thus, how long will a vaccine-mediated immune response be sustained and at what magnitude?
HOW WILL SARS-COV-2 MUTATE?
The genome of coronavirus is highly susceptible to mutations that result in genetic drift and evade immune recognition. Several studies have described this phenomenon. The genetic analysis of 86 complete or near-complete genomes of SARS-CoV-2 disclosed many mutations and deletions in coding and non-coding regions (Phan, 2020). High-resolution mapping of the SARS-CoV-2 transcriptome and epitranscriptome found at least 41 potential RNA modification sites with an AAGAA motif (Kim et al., 2020). A study of 95 complete genome sequences found 116 mutations including the three most common mutations, i.e., 8782C>T in ORF1ab, 28144T>C in ORF8, and 29095C>T in the N gene (Khailany et al., 2020). Mutations are also found in the S protein region, the crucial part for binding to human receptor ACE2. Another study reported that five of the six receptor binding domain residues of the S protein of SARS-CoV-2 differ from SARS-CoV (Andersen et al., 2020). However, this transformation does not stop there. A study identified 13 mutations in the S protein especially in spike D614G, which began to spread in Europe in early February 2020. This study also showed the evidence of recombination between the locally circulated strains indicating the multiple strain infections. (Korber et al., 2020). Twelve distinct variants were identified within the B-cell epitopes of the S protein, N protein, and M protein, and 21 distinct variants within T-cell epitopes. Of the 12 variants in the B-cell epitopes, 23403A>G Variant (p. D614 G) in an S-protein epitope has frequently been found in European countries such as the Netherlands, Switzerland and France, but rarely seen in China (Koyama et al., 2020). However, SARS-CoV-19 might not be evolving as rapidly as other RNA viruses, but we still need much more scientific evidence. Nevertheless, rapidly evolving viruses such as influenza need to be monitored to recommend new vaccine formulations twice each year (Gerdil, 2003). Similarly, no human immunodeficiency virus vaccines exist yet (Andrews and Rowland-Jones, 2017). In these circumstances, will the genetic stability of the SARS-CoV-19 remain such an extent that let the scientists develop a safe and effective vaccine?
ARE WE PREPARED FOR VACCINE BACKFIRES?
In this pressing time, perhaps some drug makers will rush through small-scale human tests that might not provide sufficient scrutiny of side effects or backfires. However, no vaccination is entirely free of any side effects or complications, and most are preventable illnesses (Kimmel, 2002). Recently, the phase 1/2 clinical trial of the ChAdOx1 nCoV-19 (NCT04324606) vaccine against COVID-19 also reported side effects such as fever, pain, muscle aches, chills, headache, and uneasiness. The research team claimed that these effects can be reduced by prophylactic paracetamol (Folegatti et al., 2020). Nevertheless, extreme caution must be taken to scrutinize backfire-effects i.e. the undesirable adverse effects (Table 2). One such dangerous backfire is vaccine-induced enhancement, which has been a major bottleneck in the development of certain corona-, flavi-, lenti-, and paramyxovirus vaccines. Here, antibody-dependent enhancement (ADE) performs a key role (Huisman et al., 2009). One study reported that the recombinant vaccinia virus Ankara expressing the S protein of SARS-CoV increased hepatitis in ferrets (Weingartl et al., 2004). Anti-S protein IgG against SARS-CoV caused severe acute lung injury in macaques (Liu et al., 2019). New Zealand white rabbits displayed increased lung inflammation after re-infection with MERS-CoV due to the lack of non-neutralizing antibodies and complement proteins (Houser et al., 2017). Researchers assume that SARS-CoV-2 severity is a consequence of ADE (Tetro, 2020). However, a study opined that the ADE and immunopathology are linked to the inflammatory feedback of host Th17, and this can be overcome by using alum as an adjuvant (Hotez et al., 2020). Now, the question arises. Will it be possible to overcome the backfires in developing COVID-19 vaccine? In brief, researchers need to resolve these questions to develop effective and safe vaccines against COVID-19 infection. However, this is not a comprehensive list as COVID-19 infection is a dynamic phenomenon depending on a lot of factors.
Table 2.
Historical concerns about vaccine safety
| Incident | Year | Consequence | Reference |
|---|---|---|---|
| Cutter Incident | 1955 | Started a polio epidemic. Two production pools accounting for 120,000 doses made by Cutter Laboratories caused 40,000 cases of polio; 51 were paralyzed, and five killed even though the vaccine had passed safety testing. | Offit, 2005 |
| Simian Virus 40 (SV40) | 1955 to 1963 | From 1955 to 1963, an estimated 10-30% of polio vaccines administered in the US were contaminated with SV40, leading to the development of a certain type of cancer. | Stratton et al., 2002 |
| Respiratory Syncytial Virus (RSV) | 1966 | Of the 20 children who underwent the FI-RSV vaccine trial, 16 needed hospitalization, two died afterwards. On contrary, only one of the 21 control group participants was hospitalized. FDA promptly suspended all clinical trials. | Kim et al., 1969 |
| H1N1 Swine Flu Vaccine and Guillain-Barré Syndrome (GBS) | 1976 | Increased risk of GBS, a rare neurological disorder. | Schonberger et al., 1979 |
| Hepatitis B Vaccine (HBV) and Multiple Sclerosis (MS) | 1998 | A relationship between HBV vaccine and MS has been suggested but is disputed. | Ascherio et al., 2001; Naismith and Cross, 2004; Le Houézec, 2014 |
| Rotavirus Vaccine and Intussusception | 1998 to 1999 | Suspension after 15 cases of intussusception, a bowel obstruction in which one segment of bowel becomes enfolded within another segment. | Iskander et al., 2004 |
| H1N1 Influenza Vaccine and Narcolepsy | 2009 to 2010 | Concern raised after abrupt-onset childhood narcolepsy was seen in Finland in 2010, but not observed in other countries including in the USA. | Partinen et al., 2012; Duffy et al., 2014 |
| Dengue Virus Vaccine- Dengvaxia | 2017 | Excess risk of severe dengue in seronegative vaccine recipients compared to seronegative non-vaccinated individuals. The Philippines stopped their immunization program after getting this warning. | Wilder-Smith, 2020 |
EFFECTIVE DEVELOPMENT STRATEGIES
The design and development of an efficacious vaccine is always a complex work, particularly in the case of SARS-CoV-2 that already have been mentioned in the previous sections. On this hurdles race to reach that endpoint of efficacious vaccine, several initiatives can be taken. First, a detailed characterization of COVID-19 immunopathogenesis should be carried out continuously, so that we can have a substantial and scientifically endorsed dataset to make the SARS-CoV-2 more predictable and understandable. Scientists and researchers have already provided several appreciable insights about the novel coronavirus, and they are working relentlessly. As this a global issue and the research is going on a global scale, global collaboration is essential to compare and validate the outcomes of the studies in a wide range of contexts and health care systems. This collaboration and annotating data could break the obstacles and enhance the probability of picking the speed of discovery up as well. Second, utilization of new technologies could overcome those stumble blocks lying in the road of COVID-19 vaccine development. Besides, conventional inactivated or live attenuated virus-vectored, new developments for non-viral vaccines, such as viral particle-like and nanostructures vaccines, subunit vaccines, RNA/DNA vaccines, and the development of rational vaccines, could provide groundbreaking approaches to addressing current vaccine production problems (Brisse et al., 2020). The new technologies for vaccine development already showing silver lining for COVID-19 vaccine, and we have noted such vaccines in the types of vaccines section of this article (Table 1). Third, the continuous development of candidate vaccines by the ongoing preclinical and clinical trials across the world. The studied population should be representative of the wide range of people based on ethnicity, habitat settings, geographic location, gender, age group, and underlying health conditions (Gaebler and Nussenzweig, 2020).
However, there may be no single winner of the vaccine; thus, it will be necessary to standardize different efficiency endpoints to permit reliable estimates and ensuring the deployment of the most successful candidates (Hodgson et al., 2020). Finally, artificial intelligence (AI) could play a significant and unprecedented role in solving problems to develop the COVID-19 vaccine. As all the probable ways discussed above need huge amounts of data to be evaluated, analyzed, and validated, AI could do the work with minimal cost, time, and effort compared to the existing setup. Besides, in silico method has already been used in developing drug and vaccine candidates. Therefore, the computational models may help us to find the candidate vaccines and therapeutics if it is feed with a sufficient amount of data (Keshavarzi Arshadi et al., 2020). In the end, in every hurdle race, sensibility, skill, and speed are indeed required to reach the finishing point successfully.
CONCLUSIONS
The number of morbidities and mortalities related to COVID-19 is increasing day by day. Global and local economies are on the verge of depression, which is exacerbating humanitarian crises across the globe. Most of the countries have imposed lockdown and stay-at-home-strategy to break the chain of the community transmission; however, these preventive methods are not sustainable for a long time. As such, there is a dire need for a vaccine against COVID-19. An efficient vaccine is the best option for controlling and prevention COVID-19 pandemic. Addressing the raised questions in this paper will improve the safety and efficacy of any COVID-19 vaccine.
ACKNOWLEDGMENTS
This research was funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF), grant number 2017R1A6A1A03015642.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- Adams E. R., Ainsworth M., Anand R., Andersson M. I., Auckland K., Baillie J. K., Barnes E., Beer S., Bell J. I., Berry T. Antibody testing for COVID-19: a report from the national COVID scientific advisory panel. Wellcome Open Res. 2020;5:139. doi: 10.12688/wellcomeopenres.15927.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., Garry R. F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. M., Rowland-Jones S. Recent advances in understanding HIV evolution. F1000Res. 2017;6:597. doi: 10.12688/f1000research.10876.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascherio A., Zhang S. M., Hernán M. A., Olek M. J., Coplan P. M., Brodovicz K., Walker A. M. Hepatitis B vaccination and the risk of multiple sclerosis. N. Engl. J. Med. 2001;344:327–332. doi: 10.1056/NEJM200102013440502. [DOI] [PubMed] [Google Scholar]
- Bernstein K. E., Khan Z., Giani J. F., Cao D. Y., Bernstein E. A., Shen X. Z. Angiotensin-converting enzyme in innate and adaptive immunity. Nat. Rev. Nephrol. 2018;14:325–336. doi: 10.1038/nrneph.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisse M., Vrba S. M., Kirk N., Liang Y., Ly H. Emerging concepts and technologies in vaccine development. Front. Immunol. 2020;11:583077. doi: 10.3389/fimmu.2020.583077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow K., Parry H., Sergeant M., Tyrrell D. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105:435–446. doi: 10.1017/S0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem A. S. Fundamentals of vaccine immunology. J. Glob. Infect. Dis. 2011;3:73–78. doi: 10.4103/0974-777X.77299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V. M., Muth D., Niemeyer D., Drosten C. Advances in Virus Research, Vol. 100. Elsevier; 2018. Hosts and sources of endemic human coronaviruses; pp. 163–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W., Fan Y., Wu W., Zhang F., Wang J. y., Ni A. p. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin. Infect. Dis. 2003;37:857–859. doi: 10.1086/378587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhu F., Guo F., Yang B., Wang T. Detection of antibodies against SARS-CoV-2 in patients with COVID-19. J. Med. Virol. 2020:doi: 10.1002/jmv.25820. doi: 10.1002/jmv.25820. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J., Weintraub E., Vellozzi C., DeStefano F. Narcolepsy and influenza A (H1N1) pandemic 2009 vaccination in the United States. Neurology. 2014;83:1823–1830. doi: 10.1212/WNL.0000000000000987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewer K., Sebastian S., Spencer A. J., Gilbert S., Hill A. V., Lambe T. Chimpanzee adenoviral vectors as vaccines for outbreak pathogens. Hum. Vaccin. Immunother. 2017;13:3020–3032. doi: 10.1080/21645515.2017.1383575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folegatti P. M., Ewer K. J., Aley P. K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E. A., Dold C., Faust S. N., Finn A., Flaxman A. L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A. M., Pollock K. M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A. D., Hill A. V. S., Lambe T., Gilbert S. C., Pollard A. J. Oxford COVID Vaccine Trial Group, author. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Nussenzweig M. C. All eyes on a hurdle race for a SARS-CoV-2 vaccine. Nature. 2020;586:501–502. doi: 10.1038/d41586-020-02926-w. [DOI] [PubMed] [Google Scholar]
- Gerdil C. The annual production cycle for influenza vaccine. Vaccine. 2003;21:1776–1779. doi: 10.1016/S0264-410X(03)00071-9. [DOI] [PubMed] [Google Scholar]
- Gopinathan U., Peacocke E., Gouglas D., Ottersen T., Røttingen J. A. Infectious Diseases in the New Millennium. Springer; 2020. R&D for emerging infectious diseases of epidemic potential: sharing risks and benefits through a new coalition; pp. 137–165. [DOI] [Google Scholar]
- Guo Y. R., Cao Q. D., Hong Z. S., Tan Y. Y., Chen S. D., Jin H. J., Tan K. S., Wang D. Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-an update on the status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson S. H., Mansatta K., Mallett G., Harris V., Emary K. R., Pollard A. J. What defines an efficacious COVID-19 vaccine? A review of the challenges assessing the clinical efficacy of vaccines against SARS-CoV-2. Lancet Infect. Dis. 2020:doi: 10.1016/S1473-3099(20)30773-8. doi: 10.1016/S1473-3099(20)30773-8. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez P. J., Corry D. B., Bottazzi M. E. COVID-19 vaccine design: the Janus face of immune enhancement. Nat. Rev. Immunol. 2020;20:347–348. doi: 10.1038/s41577-020-0323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser K. V., Broadbent A. J., Gretebeck L., Vogel L., Lamirande E. W., Sutton T., Bock K. W., Minai M., Orandle M., Moore I. N., Subbarao K. Enhanced inflammation in New Zealand white rabbits when MERS-CoV reinfection occurs in the absence of neutralizing antibody. PLoS Pathog. 2017;13:e1006565. doi: 10.1371/journal.ppat.1006565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman W., Martina B., Rimmelzwaan G., Gruters R., Osterhaus A. Vaccine-induced enhancement of viral infections. Vaccine. 2009;27:505–512. doi: 10.1016/j.vaccine.2008.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Medicine (US) Immunization Safety Review Committe, author. Immunization Safety Review: SV40 Contamination of Polio Vaccine and Cancer. In: Stratton K., Almario D. A., McCormick M. C., editors. National Academies Press (US); Washington DC: 2002. [PubMed] [Google Scholar]
- Iskander J., Haber P., Murphy T. Suspension of rotavirus vaccine after reports of intussusception--United States, 1999. MMWR Morb. Mortal. Wkly Rep. 2004;53:786–789. [PubMed] [Google Scholar]
- Jamilloux Y., Henry T., Belot A., Viel S., Fauter M., El Jammal T., Walzer T., François B., Sève P. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun. Rev. 2020:102567. doi: 10.1016/j.autrev.2020.102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouan Y., Guillon A., Gonzalez L., Perez Y., Ehrmann S., Ferreira M., Daix T., Jeannet R., Francois B., Dequin P. F., Si-Tahar M., Baranek T., Paget C. Functional alteration of innate T cells in critically ill Covid-19 patients. medRxiv. 2020:doi: 10.1101/2020.05.03.20089300. doi: 10.1101/2020.05.03.20089300. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbani N., Olds J. L. Does COVID19 infect the brain? If so, smokers might be at a higher risk. Mol. Pharmacol. 2020;97:351–353. doi: 10.1124/molpharm.120.000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarzi Arshadi A., Webb J., Salem M., Cruz E., Calad-Thomson S., Ghadirian N., Collins J., Diez-Cecilia E., Kelly B., Goodarzi H., Yuan J. S. Artificial intelligence for COVID-19 drug discovery and vaccine development. Front. Artif. Intell. 2020;3:65. doi: 10.3389/frai.2020.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khailany R. A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J. Y., Yang J. S., Kim J. W., Kim V. N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. W., Canchola J. G., Brandt C. D., Pyles G., Chanock R. M., Jensen K., Parrott R. H. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- Kimmel S. R. Vaccine adverse events: separating myth from reality. Am. Fam. Physician. 2002;66:2113–2120. [PubMed] [Google Scholar]
- Korber B., Fischer W., Gnanakaran S. G., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E. E., Bhattacharya T., Parker M. D., Partridge D. G., Evans C. M., Freeman T. M., de Silva T. I., LaBranche C. C., Montefiori D. C. on behalf of the Sheffield COVID-19 Genomics Group, author. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020:doi: 10.1016/j.cell.2020.06.043. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Weeraratne D., Snowdon J. L., Parida L. Emergence of drift variants that may affect COVID-19 vaccine development and antibody treatment. Pathogens. 2020;9:324. doi: 10.3390/pathogens9050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Houézec D. Evolution of multiple sclerosis in France since the beginning of hepatitis B vaccination. Immunol. Res. 2014;60:219–225. doi: 10.1007/s12026-014-8574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T. T., Cramer J. P., Chen R., Mayhew S. Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:667–668. doi: 10.1038/d41573-020-00151-8. [DOI] [PubMed] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., Wu T., Cheung K. W., Chan K. H., Alvarez X., Qin C., Lackner A., Perlman S., Yuen K. Y., Chen Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q. X., Liu B. Z., Deng H. J., Wu G. C., Deng K., Chen Y. K., Liao P., Qiu J. F., Lin Y., Cai X. F., Wang D. Q., Hu Y., Ren J. H., Tang N., Xu Y. Y., Yu L. H., Mo Z., Gong F., Zhang X. L., Tian W. G., Hu L., Zhang X. X., Xiang J. L., Du H. X., Liu H. W., Lang C. H., Luo X. H., Wu S. B., Cui X. P., Zhou Z., Zhu M. M., Wang J., Xue C. J., Li X. F., Wang L., Li Z. J., Wang K., Niu C. C., Yang Q. J., Tang X. J., Zhang Y., Liu X. M., Li J. J., Zhang D. C., Zhang F., Liu P., Yuan J., Li Q., Hu J. L., Chen J., Huang A. L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020a;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Long Q. X., Tang X. J., Shi Q. L., Li Q., Deng H. J., Yuan J., Hu J. L., Xu W., Zhang Y., Lv F. J., Su K., Zhang F., Gong J., Wu B., Liu X. M., Li J. J., Qiu J. F., Chen J., Huang A. L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020b;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Morris S. J., Sebastian S., Spencer A. J., Gilbert S. C. Simian adenoviruses as vaccine vectors. Fut. Virol. 2016;11:649–659. doi: 10.2217/fvl-2016-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M., Leo O. Key concepts in immunology. Vaccine. 2010;28:C2–C13. doi: 10.1016/j.vaccine.2010.07.022. [DOI] [PubMed] [Google Scholar]
- Mullard A. COVID-19 vaccine development pipeline gears up. Lancet. 2020;395:1751–1752. doi: 10.1016/S0140-6736(20)31252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith R. T., Cross A. H. Does the hepatitis B vaccine cause multiple sclerosis? Neurology. 2004;63:772–773. doi: 10.1212/01.WNL.0000137887.24504.30. [DOI] [PubMed] [Google Scholar]
- Offit P. A. The Cutter incident, 50 years later. N. Engl. J. Med. 2005;352:1411–1412. doi: 10.1056/NEJMp048180. [DOI] [PubMed] [Google Scholar]
- Partinen M., Saarenpää-Heikkilä O., Ilveskoski I., Hublin C., Linna M., Olsén P., Nokelainen P., Alén R., Wallden T., Espo M., Rusanen H., Olme J., Sätilä H., Arikka H., Kaipainen P., Julkunen I., Kirjavainen T. Increased incidence and clinical picture of childhood narcolepsy following the 2009 H1N1 pandemic vaccination campaign in Finland. PLoS ONE. 2012;7:e33723. doi: 10.1371/journal.pone.0033723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeri N. C., Shrestha N., Rahman M. S., Zaki R., Tan Z., Bibi S., Baghbanzadeh M., Aghamohammadi N., Zhang W., Haque U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int. J. Epidemiol. 2020;49:717–726. doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratan Z. A., Hosseinzadeh H., Runa N. J., Uddin B. M. M., Haidere M. F., Sarker S. K., Zaman S. B. Novel Coronavirus: a new challenge for medical scientist? Bangladesh J. Infect. Dis. 2020;7:S58–S60. doi: 10.3329/bjid.v7i0.46805. [DOI] [Google Scholar]
- Robbiani D. F., Gaebler C., Muecksch F., Lorenzi J. C. C., Wang Z., Cho A., Agudelo M., Barnes C. O., Gazumyan A., Finkin S., Hagglof T., Oliveira T. Y., Viant C., Hurley A., Hoffmann H. H., Millard K. G., Kost R. G., Cipolla M., Gordon K., Bianchini F., Chen S. T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A. W., Waltari E., Pak J. E., Huey-Tubman K. E., Koranda N., Hoffman P. R., West A. P., Jr., Rice C. M., Hatziioannou T., Bjorkman P. J., Bieniasz P. D., Caskey M., Nussenzweig M. C. Convergent antibody responses to SARS-CoV-2 infection in convalescent individuals. bioRxiv. 2020:doi: 10.1101/2020.05.13.092619. doi: 10.1101/2020.05.13.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R. A., Ferreira A. J., Simões E Silva A. C. Recent advances in the angiotensin-converting enzyme 2-angiotensin(1-7)-Mas axis. Exp. Physiol. 2008;93:519–527. doi: 10.1113/expphysiol.2008.042002. [DOI] [PubMed] [Google Scholar]
- Schonberger L. B., Bregman D. J., Sullivan-Bolyai J. Z., Keenlyside R. A., Ziegler D. W., Retailliau H. F., Eddins D. L., Bryan J. A. Guillain-Barré syndrome following vaccination in the national influenza immunization program, United States, 1976-1977. Am. J. Epidemiol. 1979;110:105–123. doi: 10.1093/oxfordjournals.aje.a112795. [DOI] [PubMed] [Google Scholar]
- Shereen M. A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetro J. A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Inf. 2020;22:72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D., de Wit E. Next-generation vaccine platforms for COVID-19. Nat. Mater. 2020;19:810–812. doi: 10.1038/s41563-020-0746-0. [DOI] [PubMed] [Google Scholar]
- Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., Godbout K., Parsons T., Baronas E., Hsieh F., Acton S., Patane M., Nichols A., Tummino P. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J., Smith G., Jones S., Proulx R, Deschambault Y., Grudeski E., Andonov A., He R., Li Y., Copps J., Grolla A., Dick D., Berry J., Ganske S., Manning L., Cao J. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol. 2004;78:12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder-Smith A. Dengue vaccine development: status and future. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2020;63:40–44. doi: 10.1007/s00103-019-03060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, author. Coronavirus Disease (COVID-19) Situation Reports, Vol. 2020. World Health Organization: WHO; 2020. Coronavirus disease (COVID-19) weekly epidemiological update and weekly operational update. [Google Scholar]
- Xiao A. T., Gao C., Zhang S. Profile of specific antibodies to SARS-CoV-2: the first report. J. Infect. 2020;81:147–178. doi: 10.1016/j.jinf.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanpanah F., Hamblin M. R., Rezaei N. The immune system and COVID-19: friend or foe? Life Sci. 2020;256:117900. doi: 10.1016/j.lfs.2020.117900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp F. Principles of vaccine design-lessons from nature. Vaccine. 2010;28:C14–C24. doi: 10.1016/j.vaccine.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhao Y. X., Zhang Y. H., Zhu L., Deng B. P., Zhou Z. L., Li S. Y., Lu X. T., Song L. L., Lei X. M., Tang W. B., Wang N., Pan C. M., Song H. D., Liu C. X., Dong B., Zhang Y., Cao Y. Angiotensin-converting enzyme 2 attenuates atherosclerotic lesions by targeting vascular cells. Proc. Natil. Acad. Sci. U.S.A. 2010;107:15886–15891. doi: 10.1073/pnas.1001253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Yang J., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W. China Novel Coronavirus Investigating and Research Team, author. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]