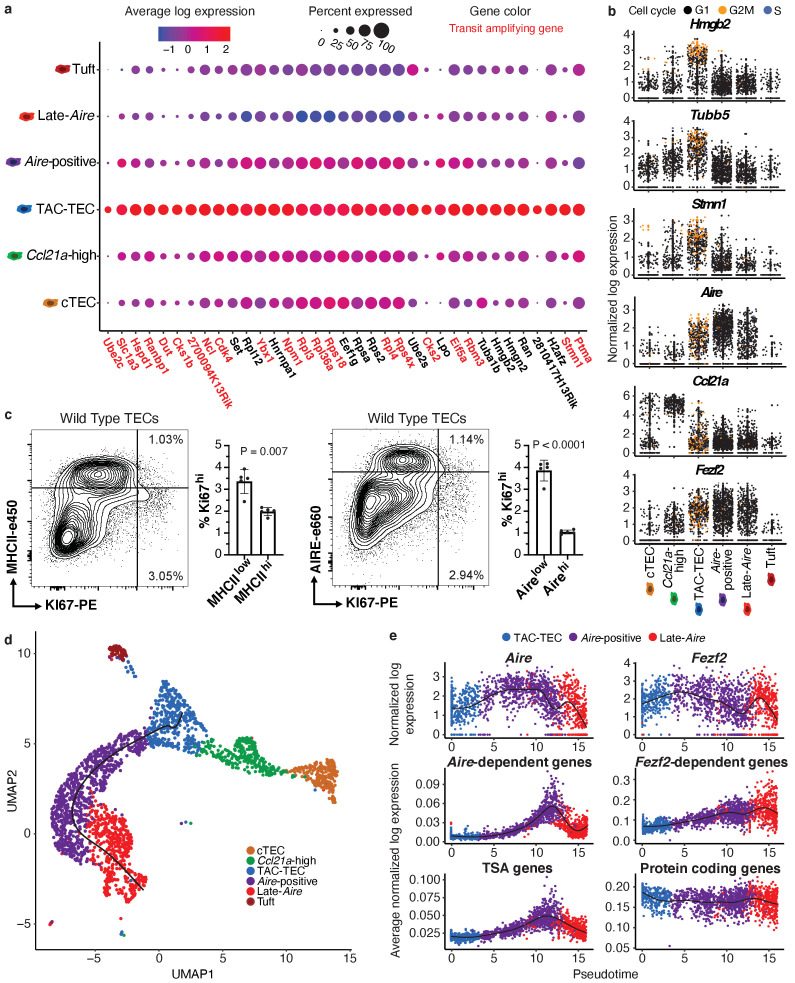
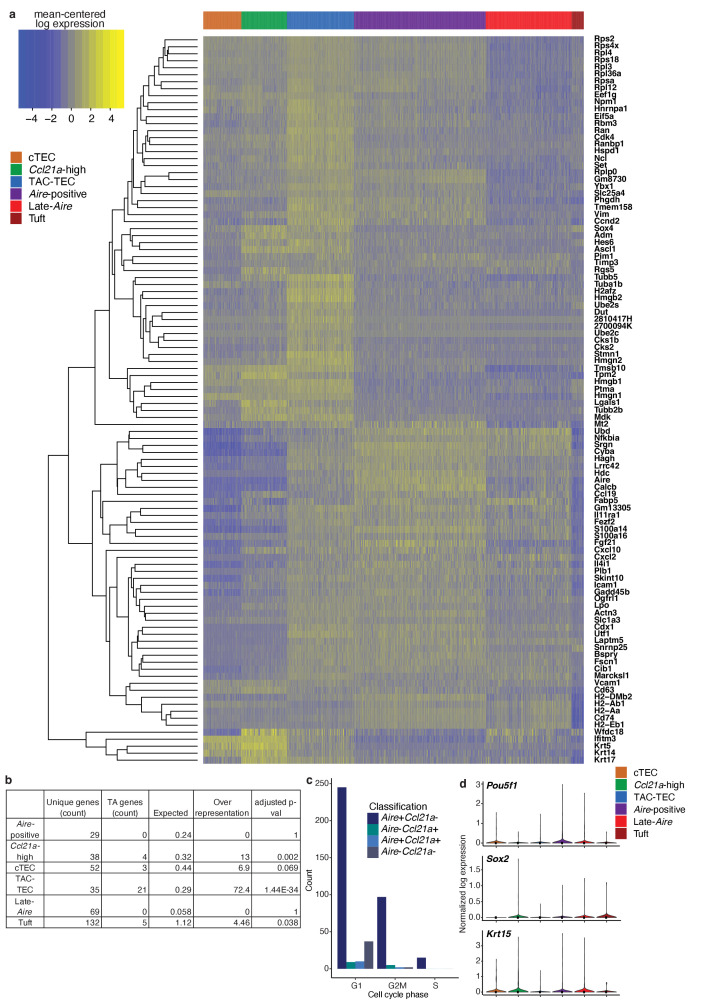
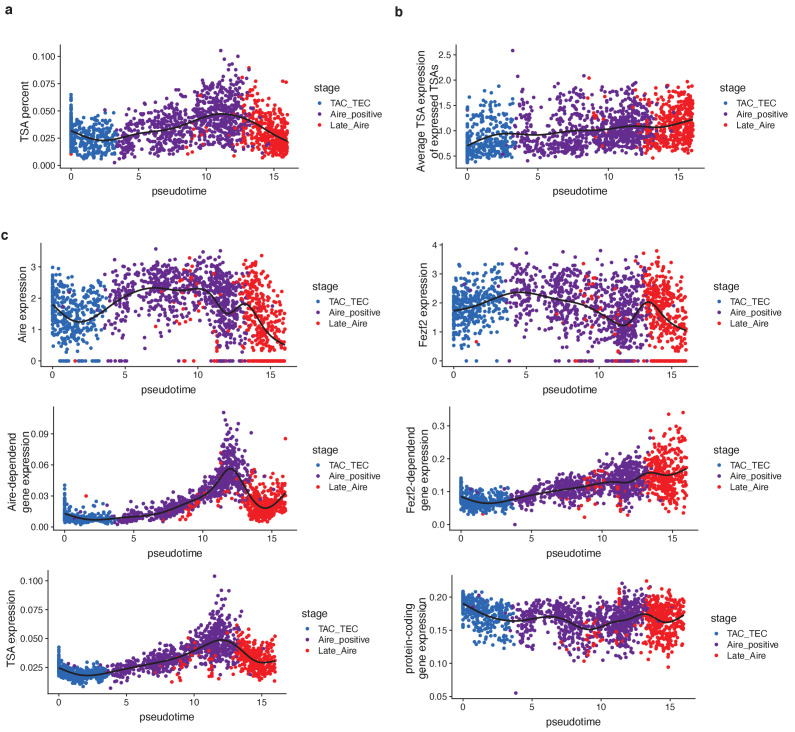
Figure 2. Characterization of TAC-TECs and developmental dynamics from this population.
(A) Summary dot plot of expression of genes differentially upregulated uniquely in the TAC-TEC population (adj-p-val<0.05 and log|fold change| > 2) across mTEC populations. Red text color represents genes previously associated with transit-amplifying cells, DE genes were enriched for transit-amplifying genes (p=1.44×10-34). Color indicates average expression and size indicates percent of cells expressing the transcript. (B) Normalized log expression of selected cell-cycle genes, chromatin modifiers, and genes important for mTEC function. Colors represent cell-cycle state. (C) Flow-cytometric analysis of mTEC (AIRE and MHCII) vs a cell-cycle (KI67) markers (three pooled thymi per replicate, n = 5 replicates). Bar plots represent percentages of cells positive for Ki67 for each population (data are mean ±s.d. n = 5 mice). (D) Predicted pseudotime line (slingshot algorithm) depicting developmental trajectory exclusively for cells in the Aire branch. (E) Expression patterns of genes (log expression) and gene sets (average log expression) for cells in the Aire branch of development across pseudotime (see E). Color represents population. See also Figure 2—figure supplements 1 and 2.