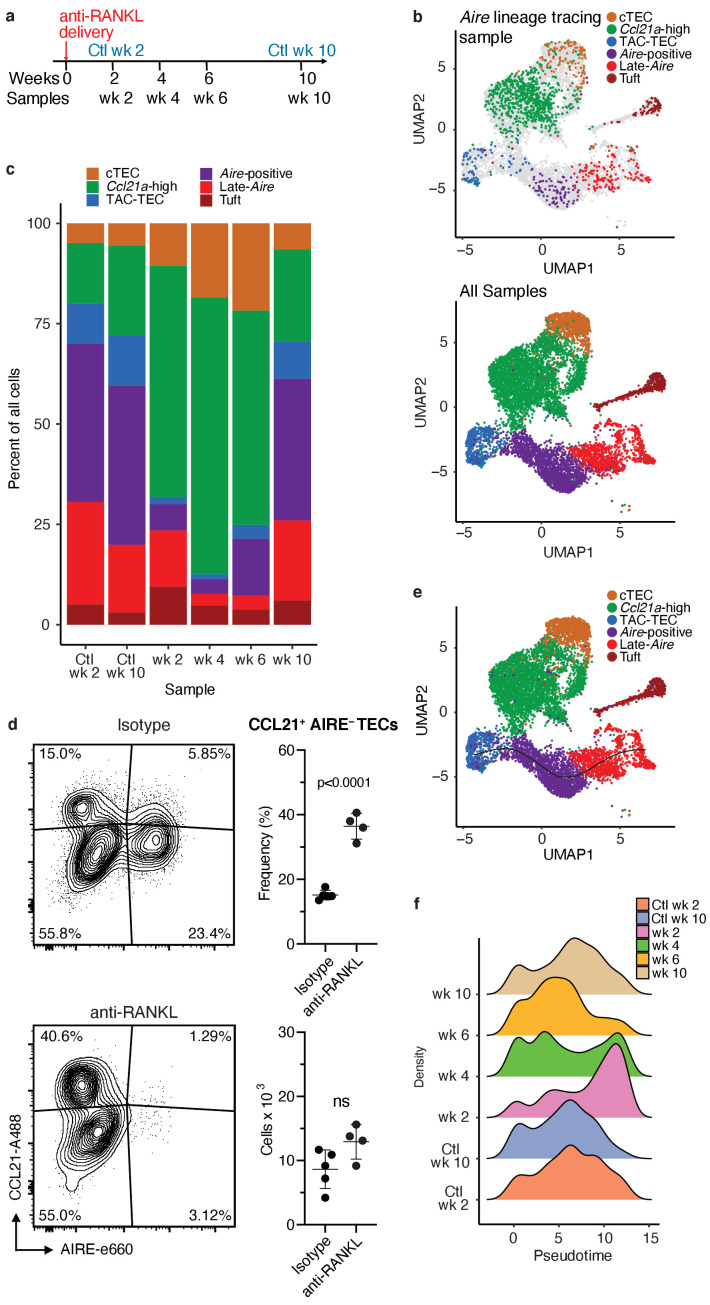
Figure 4. Treatment with anti-RANK ligand decreases the relative size of the entire Aire-expressing mTEC population.
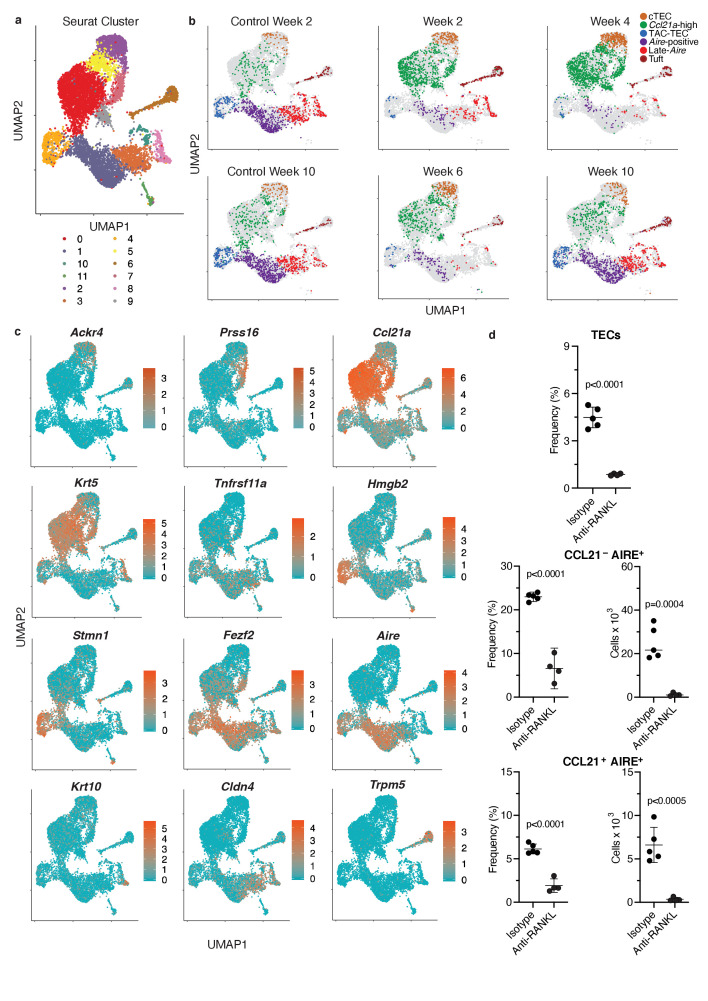
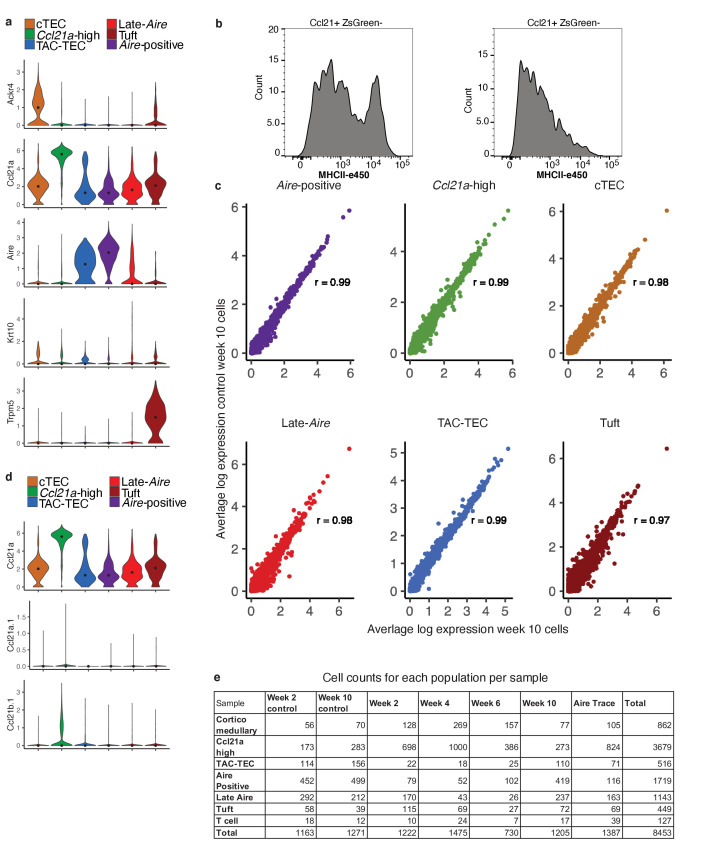
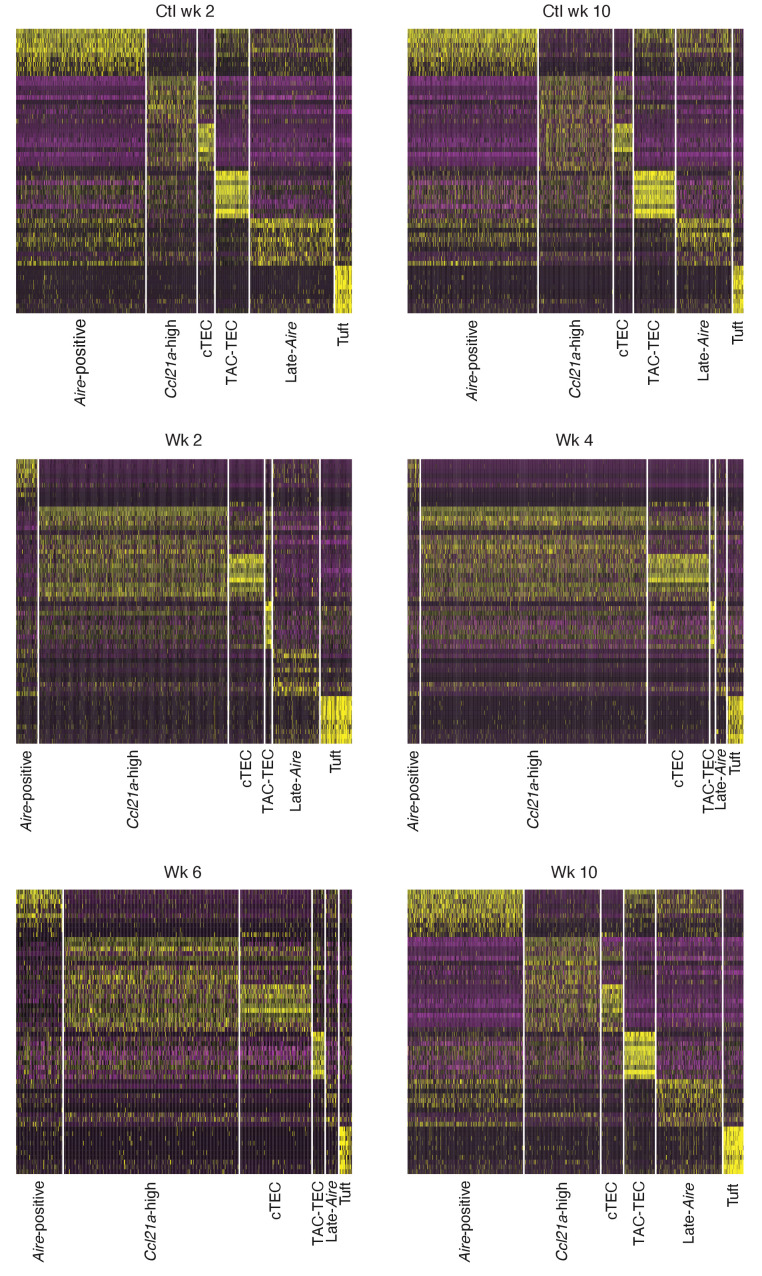
(A) Overview of the experimental protocol. Anti-RANKL was given over the course of a week, and thymi were sequenced at weeks 2, 4, 6, and 10 following treatment. Isotype control thymi were also sequenced at weeks 2 and 10. 3 thymi were pooled for all samples. (B) UMAP projections of all 8453 cells from all samples. Top: color represents original Aire trace identity (see Figure 1), gray represents all other samples. Bottom: color represents inferred population labels. Identity of cells in all samples was inferred from the original identity of Aire lineage tracing cells. (C) Proportions of cells in each population for each sample. Color is cell population. (D) Flow-cytometric analysis of the Ccl21a population at week four following treatment with anti-RANKL and an age-matched isotype control mouse (two pooled thymi per replicate, n = 4 replicates for treatment, n = 5 for controls). Plots show the proportion of all TECs in the CCL21 population and the absolute number of cells in the CCL21 population for the anti-RANKL treated and isotype control samples (data are mean ± s.d.). (E) Predicted pseudotime line (slingshot algorithm) depicting developmental trajectory for cells from all samples in exclusively the Aire branch. (F) Density plots of cells in the Aire branch of development across pseudotime (see E) for each sample. Color represents sample. See also Figure 4—figure supplements 1, 2 and 3.