Figure 2.
The DG Discriminates between Small Changes in Contextual Information
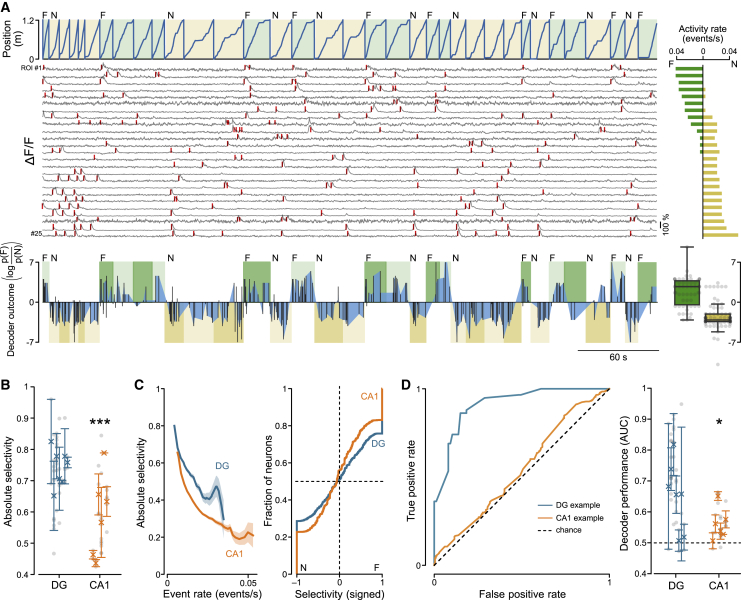
(A) Top: example session recorded from the DG, showing the position of the animal against time, the context type for each lap (F, vertical gratings in green; N, oblique gratings in yellow), and fluorescence traces extracted from the 25 most active neurons (gray), with significant transients during running periods indicated by red tick marks. For each fluorescence trace on the left, the corresponding event rate in the F (green) and N (yellow) environments is reported on the right. Bottom: decoder outcome for the session shown above. Light and dark color shades refer to training and test data, respectively (same color code as above). Black bars connected by blue areas represent the decoder outcome (difference in log likelihood of the two environments; F, positive; N, negative). Bottom right: summary of decoder outcome for the test data of the example session. Data points represent time bins.
(B) Absolute selectivity (STAR Methods) for the two environments F and N in DG (blue) and CA1 (orange) (DG, 0.74 ± 0.02; n = 7 mice; CA1, 0.59 ± 0.05; n = 6 mice; LMM, p = 0.0007). Symbols with error bars represent mean ± SEM of individual animals. Gray circles represent recording sessions.
(C) Left: absolute selectivity for DG and CA1 neurons plotted against their firing rate. Right: cumulative histogram; each neuron was scored depending on its selectivity for the F (positive) or N (negative) environment (DG, n = 4,667 active cells; CA1, n = 10,587 active cells).
(D) Left: example quantification of decoder performance (receiver operating characteristic [ROC]). Blue trace, DG session; orange trace, CA1 session. Right: comparison of decoder performance, quantified as area under the ROC curve (AUC), between the DG and CA1 (DG, 0.65 ± 0.04, n = 7 mice; CA1, 0.56 ± 0.02, n = 6 mice; LMM, p = 0.04; chance level, 0.5). Same symbols as in (B).
∗p < 0.05, ∗∗∗p < 0.001. See also Figures S1G–S1L.