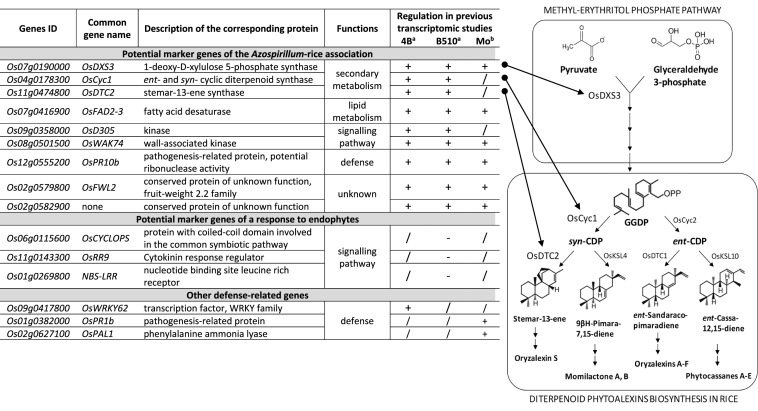
Table 3.
Description of the 15 genes selected for gene expression analysis in rice roots
The right panel displays secondary metabolism pathways in which three selected genes are involved: the Methyl-Erythritol Phosphate pathway followed by the diterpenoid phytoalexins biosynthesis in rice. GGDP geranylgeranyl diphosphate, CDP copalyl diphosphate
aTranscriptional regulation based on microarray analysis of Nipponbare roots 7 days post-inoculation with A. lipoferum 4B or Azospirillum sp. B510 (Drogue et al. 2014)
bTranscriptional regulation based on microarray analysis of Nipponbare roots 6 days post-infection with the fungal pathogen Magnaporthe oryzae (Marcel et al. 2010)
For a and b: + means up-regulation, − means down-regulation, /, means no significant regulation