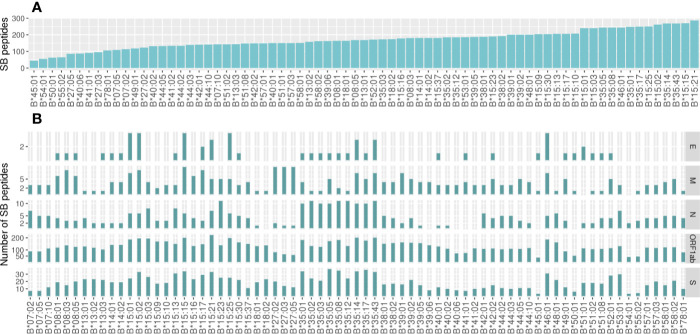
Figure 3.
Number of unique Strong Binder peptides for the 73 analyzed HLA-B alleles. (A) Global view of the predicted binding spectra for HLA-A considering the entire SARS-CoV-2 genome or (B) depicted by ORF1ab, Envelope (E), Membrane (M), Nucleocapsid (N), or Spike (S) proteins. Strong Binder peptides were selected using a %Rank cut-off of 0.5%, which is based on the likelihood of this peptide being presented when compared to a pool of natural ligands. HLA binding predictions performed by netMHCpan were crossed with TAP and proteasome predictions performed by netCTLpan in order to get a more reliable set of peptides. Importantly, the same peptide can be predicted to be presented by more than one HLA.