Figure 1. Graphical workflow for targeting S1PR3 signaling and characterizing the regenerative capacity of immune modulation in muscle injuries.
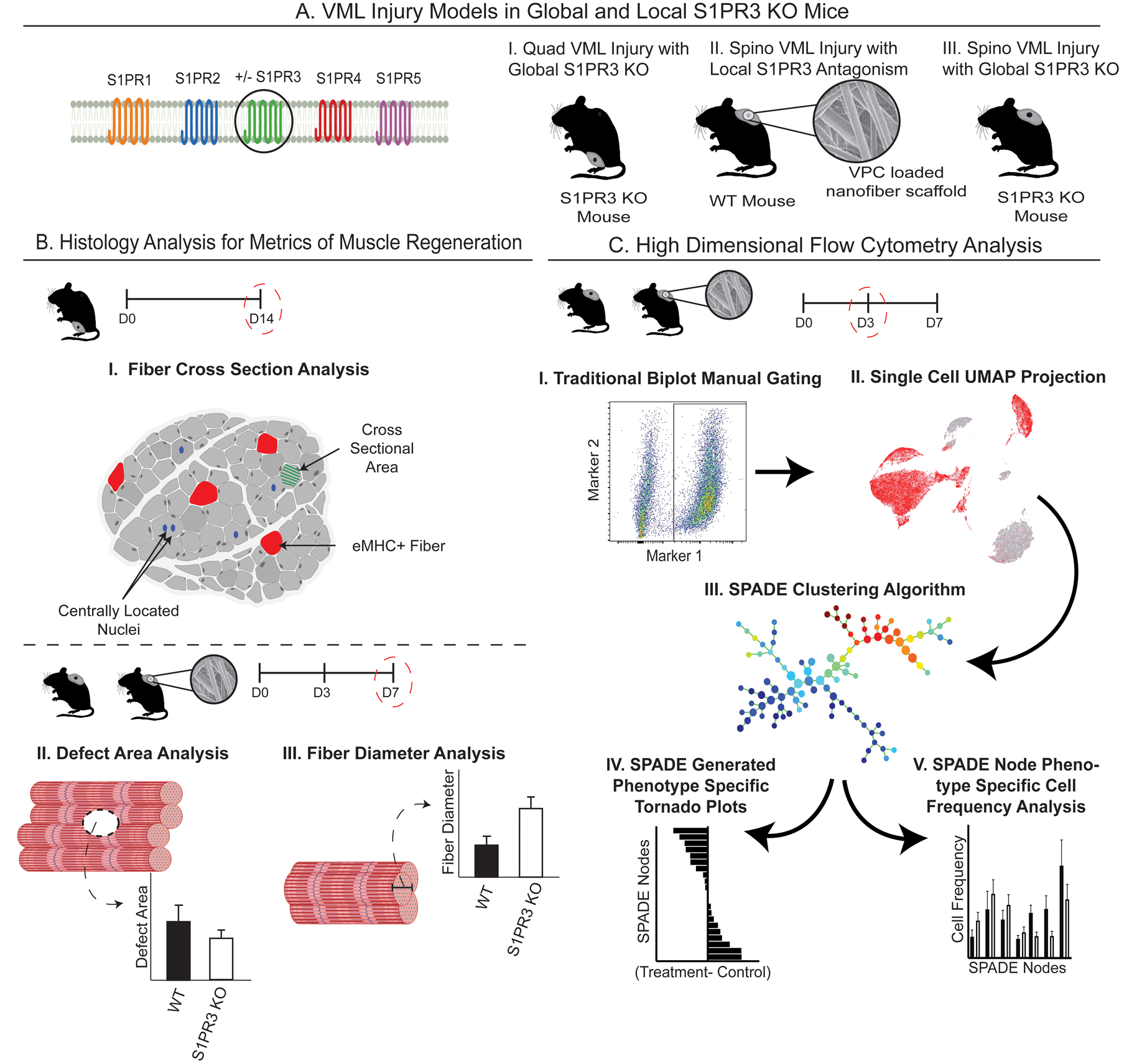
Graphical representation of the innovative methods employed in targeting of S1PR3 signaling for treatment of muscle injuries and the subsequent high dimensional analysis utilized to capture unique, infiltrating cellular subsets. (A) Introduction to the various mouse VML models used in these studies, including (A-I) quadriceps (quad) VML model in S1PR3 KO mice, (A-II) local inhibition of S1PR3 in the spinotrapezius (spino) with S1PR3 antagonist VPC01091-loaded (VPC) nanofibers in WT mice, and (A-III) a spino VML defect model in S1PR3 KO mice. (B) Metrics of muscle regeneration analyzed in these studies including histological analysis of quadricep cross sections (B-I) along with defect area and fiber diameter analysis via whole mount immunohistochemistry in the spino (B-II, III). (C) Workflow for analyzing single-cell high dimensional flow cytometry data after muscle injury. Our approach involves manually gating immune cell subsets (C-I), visualizing underlying heterogeneity via 2D projections using UMAP (C-II), high dimensional clustering algorithms with pseudotime trajectories in SPADE (C-III), and resulting tornado plots (C-IV) and SPADE node frequency quantifications (C-V). Created with BioRender.com.
