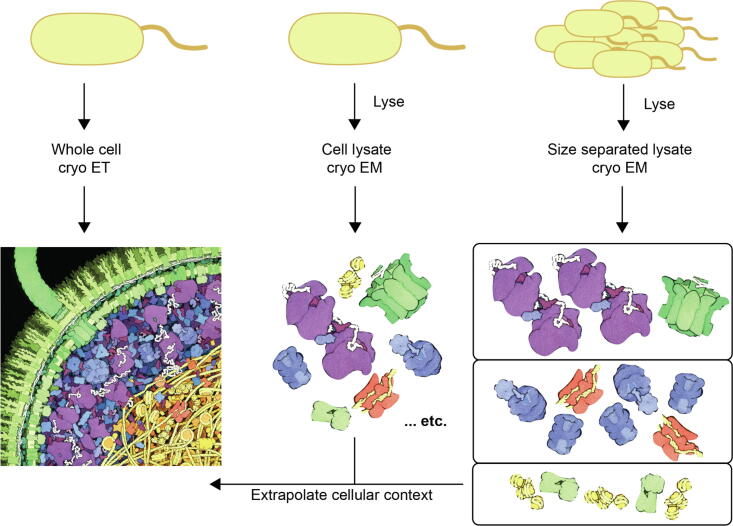
Fig. 3.
in situ structural characterization of quinary interactions. A) Cryo ET of an E. coli cell, resulting in a detailed look of the inside of the cellular interactions (illustration by David S. Goodsell, the Scripps Research Institute). B) Individual cell lysate applied directly to a cryo-EM grid results in a mixture of protein structures that have been minimally disrupted, as represented by the structures extracted directly from the cell picture. C) Separating the lysate of several cells using only size exclusion chromatography results in fractionated cryo-EM samples (represented by the particles boxed by physical size), which are computationally easier to process.