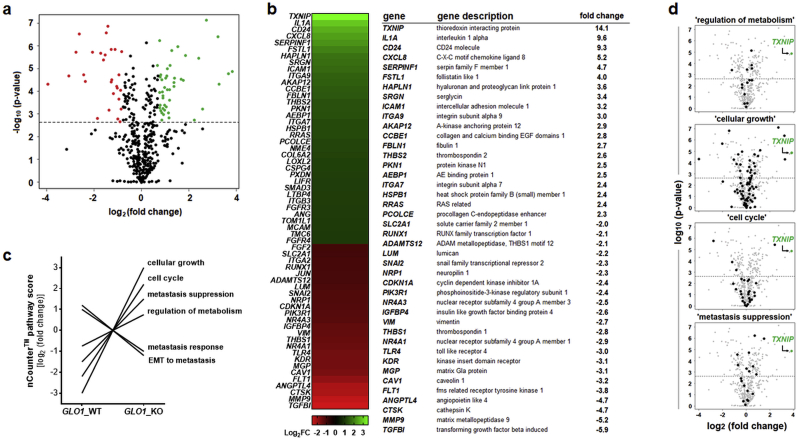
Fig. 1.
NanoString nCounter™ profiling identifies pronounced gene expression changes (including TXNIP upregulation) as a consequence of CRISPR/Cas9-based GLO1 deletion in human A375 malignant melanoma cells. NanoString™ analysis (using the nCounter™ PanCancer Progression Panel) was performed comparing gene expression between cultured human A375 malignant melanoma cells (GLO1_WT) and an isogenic variant (GLO1_KO [B40]). (a) Volcano plot [fold change (log2) versus p-value (log10)] depicting differential gene expression of 740 genes (GLO1_KO versus GLO1_WT; cut-off criteria: fold change ≥ 2; p ≤ 0.05; upregulated: green dots; downregulated: red dots). (b) Left panel: heat map depiction of statistically significant expression changes; right panel: table summarizing numerical values of up- and downregulated genes; cut off criteria as specified in (a). (c) NanoString nCounter™ covariate plot of gene expression ‘pathway scores’ as a function of GLO1 genotype identifying GLO1-responsive expression networks. (d) Volcano plots depicting individual expression pathways identified in panel (c) characterized by TXNIP upregulation representing the most pronounced expression change: ‘regulation of metabolism’ (out of 16 genes), ‘cellular growth’ (out of 97 genes), ‘cell cycle’ (out of 46 genes), and ‘metastasis suppression’ (out of 19 genes). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)