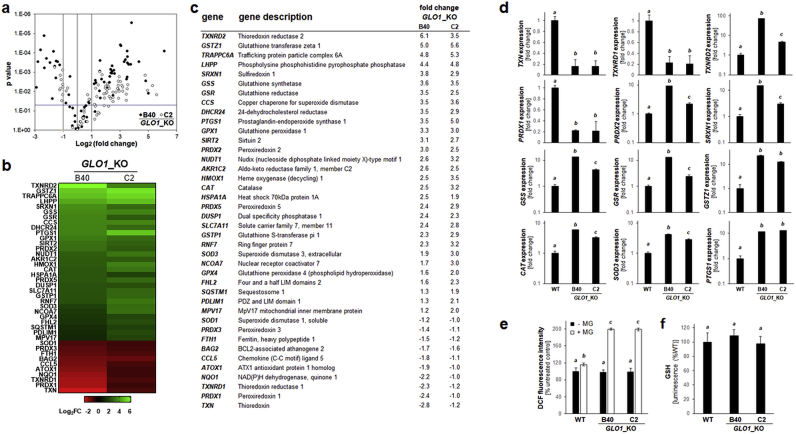
Fig. 4.
Genetic deletion of GLO1 alters redox stress response gene expression in human A375 melanoma cells. (a) RT2 Profiler™ PCR array analysis of redox stress response genes expression (GLO1_KO clones [B40 and C2] relative to GLO1_WT). Volcano plot depicts differential gene expression (cut-off criteria: expression differential ≥ 2; p value ≤ 0.05; filled circles: GLO1_KO [B40]; empty circles: GLO1_KO [C2]). (b–c) Heat map depiction of statistically significant expression changes (log2 fold change) revealing clustered modulation of redox-related genes as a function of GLO1 deletion [as summarized numerically in (c)]. (d) GLO1-modulated ‘oxidative stress response’ as revealed at the single gene expression level (GLO1_WT versus GLO1_KO clones [B40 and C2]): thioredoxin-related: TXN, TXNRD1, TXNRD2, PRDX1, PRDX2; glutathione-related: GSS, GSR, GSTZ1; other antioxidant factors: SRXN1, CAT, SOD3; inflammation: PTGS1. Bar graphs depict fold change (logarithmic or metric scale according to data range). (e) Oxidative stress (GLO1_WT versus GLO1_KO clones [B40 and C2]) as monitored by flow cytometric detection of DCF fluorescence [with or without MG treatment (500 μM, 2h)]. (f) Intracellular reduced glutathione content as assessed by luminescence intensity (GSH-Glo™) normalized to cell number (GLO1_WT versus GLO1_KO clones [B40 and C2]). For all bar graph depictions, quantitative data analysis employed ANOVA with Tukey's post hoc test; means without a common letter differ from each other (p < 0.05).