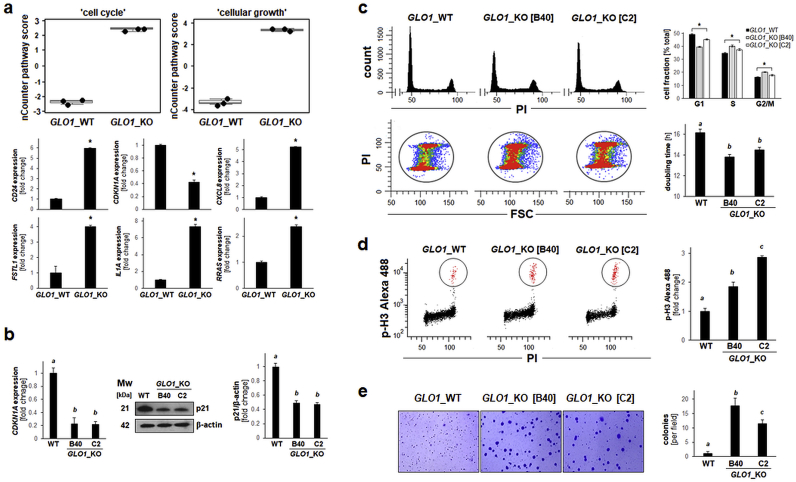
Fig. 5.
Genetic deletion of GLO1 shortens population doubling time while increasing M-phase cell cycle population and enhancing clonogenicityof human A375 melanoma cells. (a) NanoString nCounterTM pathway score analysis of ‘cell cycle’ and ‘cellular growth’ (top: box plot depiction); single panels (bottom) indicate comparative gene expression [CD24, CDKN1A, CXCL8, FSTL1, IL1A, RRAS]. (b) CDKN1A expression in A375 melanoma (GLO1_WT versus GLO1_KO clones [B40 and C2]) as confirmed by RT-qPCR (left) and immunoblot (right) analysis. Bar graph summarizes protein levels over β-actin control. (c) Alteration of cell cycle and population doubling time: Top panel (left): Representative cell cycle histograms per treatment group as assessed by flow cytometry of PI-stained cells. Top panel (right): Cell cycle distribution as summarized by bar graph depiction. Bottom panel (left): Population shift from G1-towards S- and G2/M-phases as visualized by flow cytometric analysis [PI versus FSC (forward scatter); representative images]. Bottom panel (right): Population doubling time (hr) as determined by proliferation analysis summarized by bar graph depiction. (d) M-phase population assessment as measured by flow cytometry of phospho-histone H3 (Ser10) versus PI-double-stained cells (left panels: representative images); right panel: summary of numerical results as a function of GLO1_KO status. (e) Colony formation was assessed by determining anchorage-independent growth in soft agar; representative images after crystal violet staining (left panels) as summarized by bar graph depiction (right panel). For all bar graph depictions, quantitative data analysis employed ANOVA with Tukey's post hoc test; means without a common letter differ from each other (p < 0.05). For bar graphs comparing two groups only, statistical significance was calculated employing the Student's two-tailed t-test (*p < 0.05). . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)