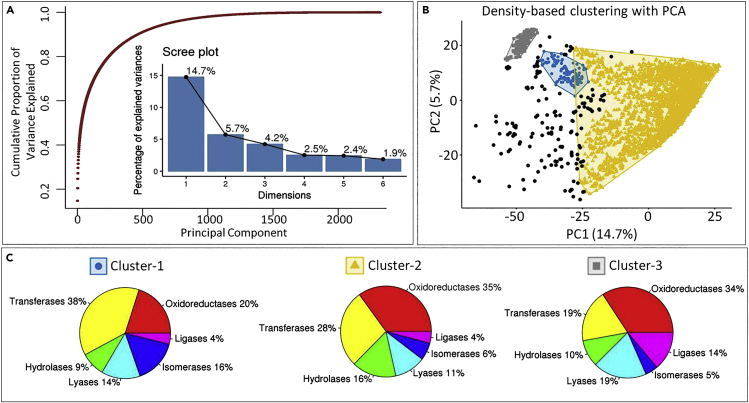
Figure 2.
Evaluating the diversity and complexity of metabolic dataset
(A) The cumulative scree plot and a normal scree plot from the PCA analysis of all the substrate molecules from the complete input dataset based on the 2,322 selected features. The x axis is the principal component number, y axis for the dot plot is the cumulative variance explained by the individual principal components, and the y axis for the bar plot is the percentage of variance explained by the individual principal components.
(B) The density-based clustering of substrate molecules using the principal component PC1 and PC2 from the PCA analysis. The density-based clustering was performed with MinPts value 20 and epsilon value 3.25. Three dense clusters were identified, which are colored as blue for cluster-1, yellow for cluster-2, and gray for cluster-3.
(C) The proportion of different reaction classes in each cluster is shown as pie chart with percentage value labeled for each reaction class. The proportion is calculated based on the number of substrate molecules that can undergo a particular kind of reaction class in that cluster. The pie chart is mentioned for all three above identified clusters: cluster-1, cluster-2, and cluster-3.
See also Figures S5 and S6.