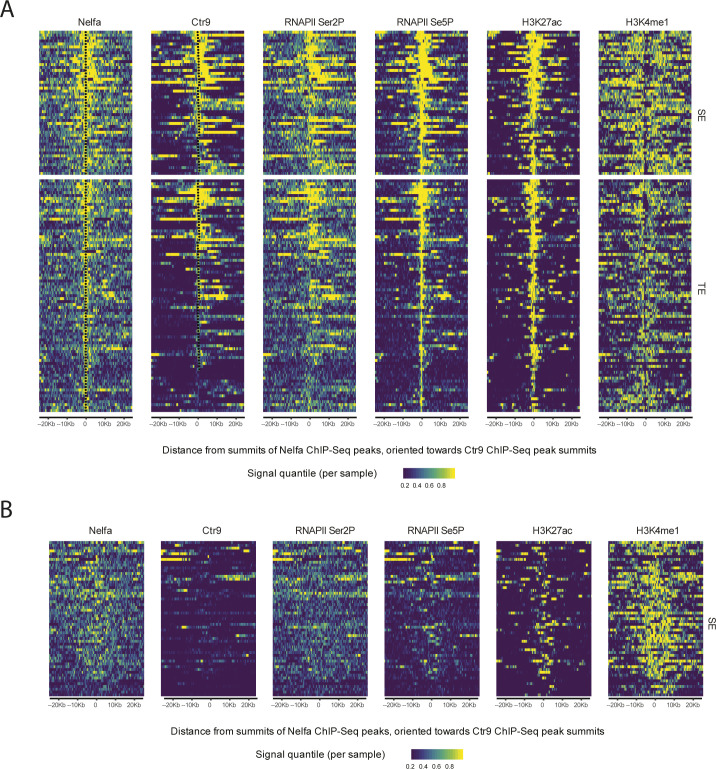
Figure S5. Zoom out view occupancy of NELFA and Ctr9 at enhancers.
(A) Heat maps showing ChIP-Seq occupancy patterns of NELFA, Ctr9, RNAPII Ser2P, RNAPII Ser5P, H3K27ac, and H3K4me1 at enhancers that are bound by NELFA. Color-coding is based on quantile-normalized read coverage signals, with yellow indicating stronger binding. Black dots covering NELFA and Ctr9 heat maps are placed at summits of predicted ChIP-Seq peaks. (B) Heat maps showing ChIP-Seq occupancy patterns of NELFA, Ctr9, RNAPII Ser2P, RNAPII Ser5P, H3K27ac, and H3K4me1 at super enhancers that are not bound by Ctr9. Each ChIP-Seq experiment was performed with a single sample. Color-coding is based on quantile-normalized read coverage signals, with yellow indicating stronger binding. All heat maps are showing 50-kb windows centered around NELFA peak summits, oriented towards the nearest Ctr9 peak summit.