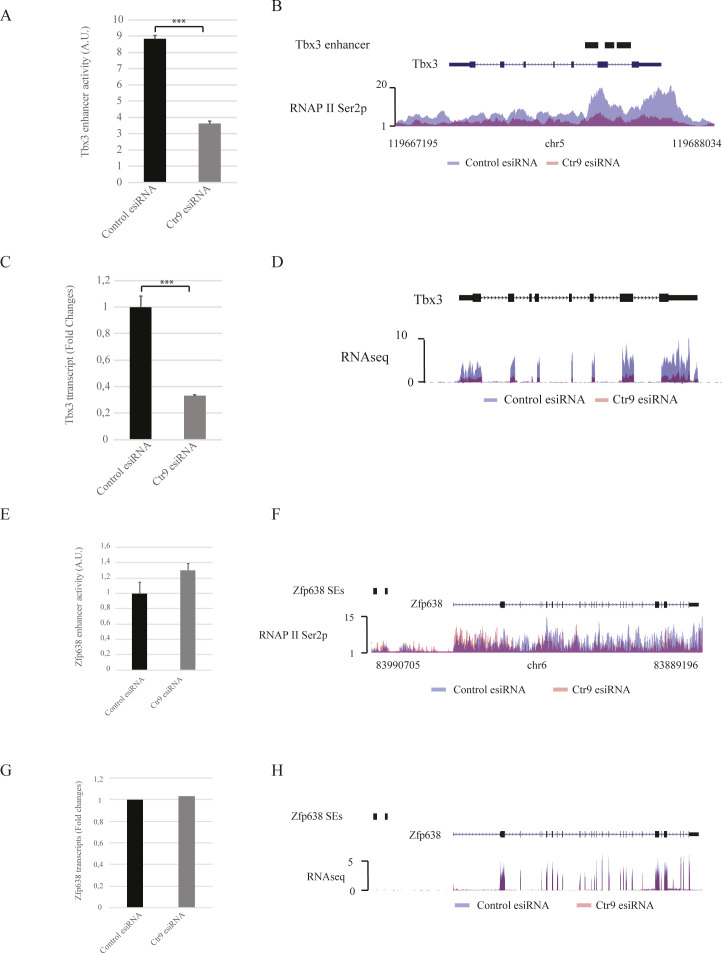
Figure S6. Paf1C regulates gene expression by modulating enhancer activity.
(A) Enhancer activity of Tbx3 super enhancer (SE) after transfection with a non-targeting control silencing trigger (black) and Ctr9 knockdown (grey). The y-axis shows the luciferase measurements in A.U. The values are normalized to sample transfected with the empty reporter vector. The pRL-SV40 plasmid was used as transfection efficiency control. Data are presented as the mean ± SD from three independent experiments. Error bars depict SD. (B) RNAPII Ser2p occupancy on Tbx3 gene after transfection with a non-targeting control silencing trigger (blue) and Ctr9 knockdown (orange). The x-axis indicates the chromosome position, and the y-axis represents normalized read density in reads per million. Black boxes indicate the annotated Tbx3 SE. Each ChIP-Seq experiment was performed with a single sample. (C) qRT-PCR quantification of Tbx3 expression after transfection with a non-targeting silencing trigger control (black) and Ctr9 knockdown (grey). Tbx3 expression was normalized to the expression of the housekeeping gene GAPDH, and shown as fold changes to the sample transfected with the non-targeting silencing trigger. Data are presented as the mean ± SD from three independent experiments. Error bars depict SD. (D) Tbx3 expression determined by RNA-seq after transfection with a non-targeting silencing trigger (blue) and Ctr9 knockdown (orange). The y-axis represents normalized read density in reads per 10 million. (E) Enhancer activity of Zfp638 SE, which is not bound by Ctr9, after transfection with a non-targeting control silencing trigger (black) and Ctr9 knockdown (grey). The y-axis shows the luciferase measurements in A.U. The values are normalized to sample transfected with the empty reporter vector. The pRL-SV40 plasmid was used as transfection efficiency control. Data are presented as the mean ± SD from three independent experiments. Error bars depict SD. (F) RNAPII Ser2p occupancy on Zfp638 gene after transfection with a non-targeting control silencing trigger (blue) and Ctr9 knockdown (orange). The x-axis indicates the chromosome position, and the y-axis represents normalized read density in reads per million. Black boxes indicate the annotated Tbx3 SE. (G) Quantification of Zfp638 expression after transfection with a non-targeting silencing trigger control (black) and Ctr9 knockdown (grey). Zfp638 expression was determined by reads from RNAseqs and shown as fold changes to the sample transfected with the non-targeting silencing trigger. (H) Zfp638 expression determined by RNA-seq after transfection with a non-targeting silencing trigger (blue) and Ctr9 knockdown (orange). The y-axis represents normalized read density in reads per 10 million. Statistically significant differences were determined by a two-tailed t test (** indicates P < 0.01 and *** indicates P < 0.001).