Figure 2. SRSF6 specifically recognizes GAA motifs in exons.
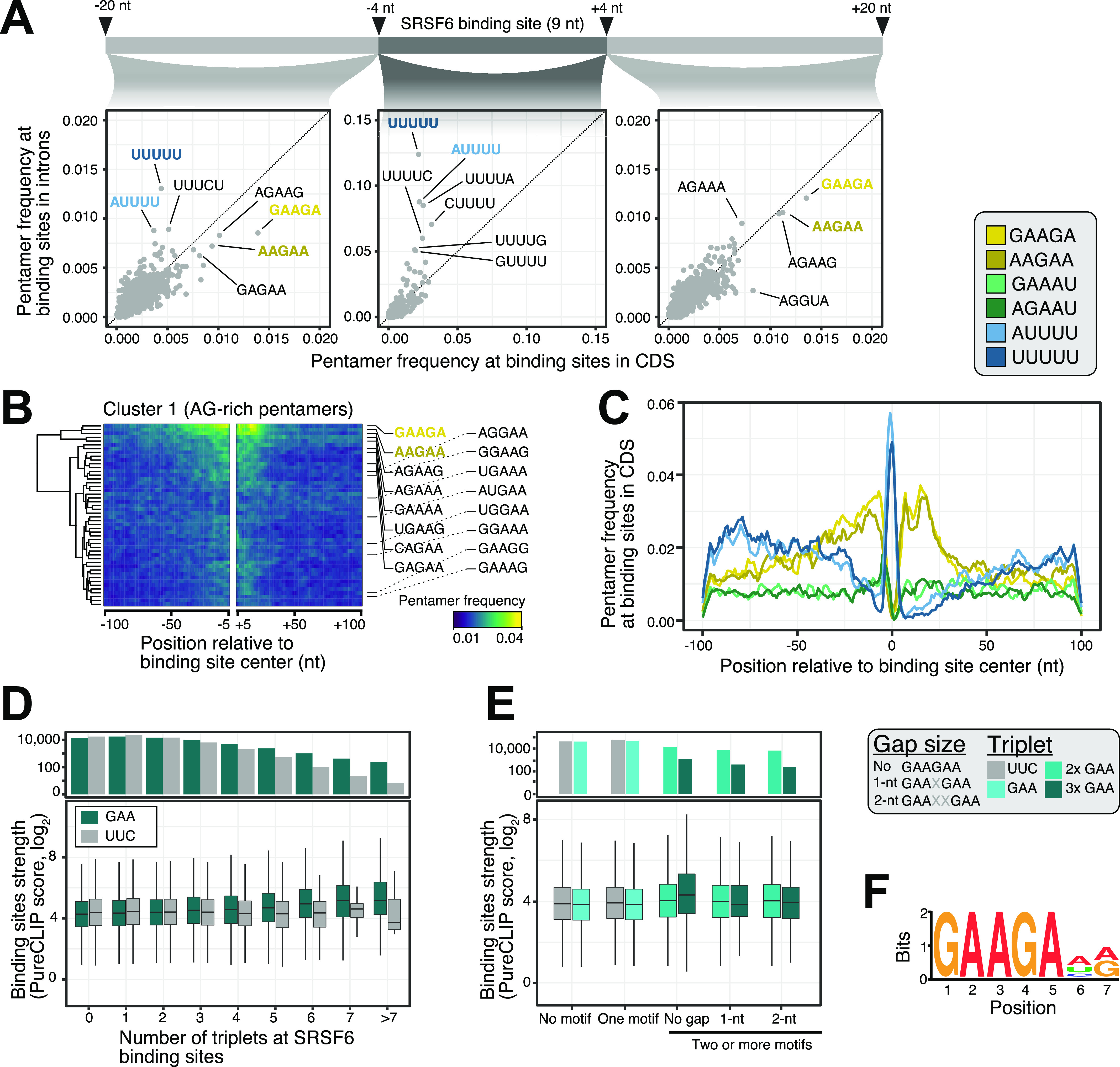
(A) SRSF6 binding sites in CDS frequently display AG-rich pentamers, contrasting the uridine-rich pentamers in introns, most prominently UUUUU. Scatter plots compare pentamer frequency within the 9-nt binding sites and in flanking 20-nt windows for SRSF6 binding sites in introns and CDS. Two most enriched pentamers of clusters 1 and 2 derived from hierarchical clustering of pentamer profiles are colored (see (B) and Fig S2A). (B) GAAGA and AAGAA enrich around SRSF6 binding sites in the CDS, whereas UUUUU marks binding site centers. Heat map shows cluster 1 from hierarchical clustering of pentamer profiles. Two most enriched pentamers are labeled and colored as in (A). Clusters 2 and 3 with uridine-rich and other pentamers are shown in Fig S2A and B. (C) SRSF6 positions towards end of motif-enriched stretches. Metaprofile shows pentamer frequencies in 201-nt window around SRSF6 binding sites. Two most enriched pentamers of three clusters from hierarchical clustering of pentamer profiles are shown. (D) Binding site strength increases with the number of GAA triplets. Boxplot (bottom) shows the distribution of binding site strengths (log2-transformed PureCLIP score) for binding sites with a given number of GAA triplets within 30 nt from binding site center. Reverse complement UUC was used as control. Box represents quartiles, center line denotes 50th percentile, and whiskers extend to most extreme data points within 1.5× interquartile range. The bar chart (top) gives number of binding sites in each category. (E) Two or more GAA triplets in direct sequence are associated with increased binding site strength. Boxplot shows the distribution of binding site strengths (log2-transformed PureCLIP score) for binding sites with no or one triplet (GAA or UCC) compared with two or more triplets in direct sequence or with 1-nt or 2-nt gaps. Visualization as in (D). (F) Motif enrichment analysis using DREME (Bailey, 2011) detected a purine-rich motif, reinforcing the role of GAA regions in SRSF6 binding. The motif is present at 25,148 SRSF6 binding sites.
