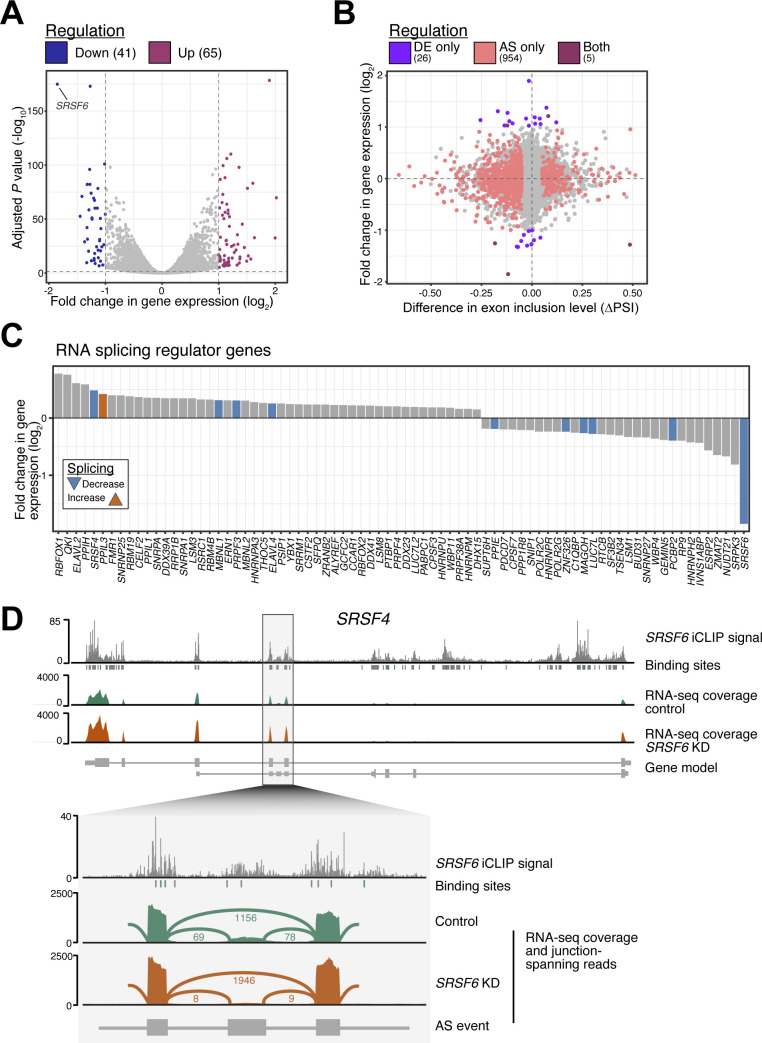
Figure S5. SRSF6 KD affects gene expression of splicing-regulatory proteins in EndoC-βH1 cells.
(A) SRSF6 KD significantly changes the expression of 106 genes. Volcano plot shows adjusted P-value (Benjamini–Hochberg correction) against log2-transformed fold-change (LFC). Significant genes are highlighted (adjusted P-value < 0.001, |LFC| > 1). (B) SRSF6-regulated splicing events are not associated with global changes in gene expression. Scatter plot compares differences in exon inclusion (percent spliced-in, PSI) to changes in gene expression upon SRSF6 KD in EndoC-βH1 cells. Genes are color-coded according to their significant differential expression and/or alternative splicing. (C) SRSF6 KD affects expression of many splicing regulator genes in EndoC-βH1 cells. The bar chart depicts log2-transformed fold-change of differentially expressed splicing regulator genes (adjusted P-value < 0.001). Genes with additional alternatively spliced exons are colored. (D) The SR protein-encoding gene SRSF4 is significantly up-regulated upon SRSF6 KD. It harbors an SRSF6-regulated exon and shows strong SRSF6 binding. Genome browser view depicts SRSF6 iCLIP data (merged replicates) and binding sites (green) together with RNA-seq read coverage from control and SRSF6 KD EndoC-βH1 cells. Selected transcript isoforms are shown below (GENCODE v29). Zoom-in on SRSF6-regulated cassette exon and the flanking constitutive exons. Lines indicate exon-exon junctions with numbers of supporting reads. In line with the global RNA splicing map for SRSF6 (Fig 3C), SRSF6 binding in the alternative exon appears to promote its inclusion.