Figure 7.
The COX-IS Predicts Response to ICB in Different Tumor Types
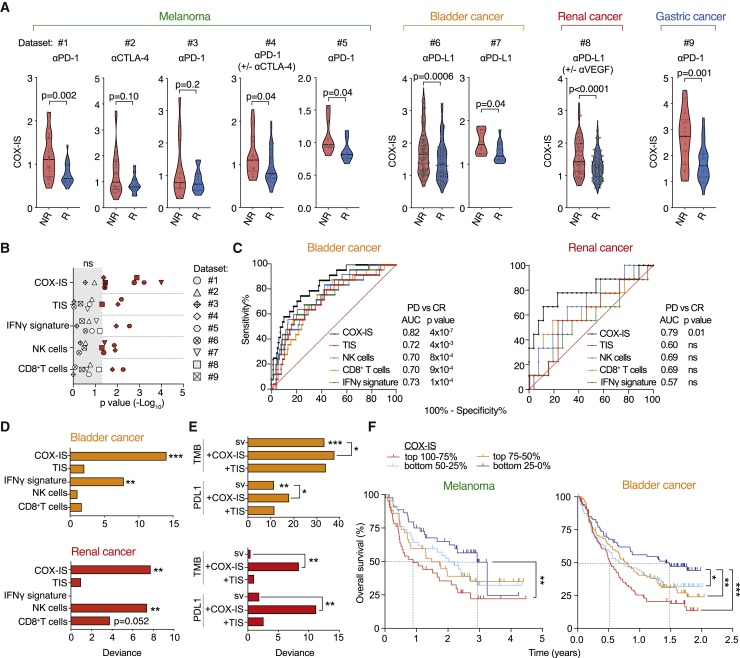
(A) Analysis of COX-IS at baseline in responder (R) and non-responder (NR) groups in melanoma (dataset 1: Riaz et al., 2017; 2: Van Allen et al., 2015; 3: Hugo et al., 2016; 4: Gide et al., 2019; 5: Chen et al., 2016), bladder (dataset 6: Mariathasan et al., 2018; 7: Snyder et al., 2017), renal (dataset 8: McDermott et al., 2018), and gastric (dataset 9: Kim et al., 2018) cancer patients as defined in the original studies (see STAR Methods).
(B) Analysis of COX-IS; TIS; and IFN-γ, NK cell, and CD8+ T cell signatures (see Table S4) at baseline in R and NR patients shown in (A). The p value (−log10) for each comparison is plotted.
(C) ROC analysis for COX-IS; TIS; and IFN-γ, NK cell, and CD8+ T cell signatures in PD versus CR patient from datasets 6 and 8. The area under the ROC curve was used to quantify response prediction.
(D and E) Explained variance (deviance) in patient response for generalized linear models fit using single variables (sv) (D) or their combinations with TMB or PD-L1 expression (E) in dataset 6 and 8. Chi-square test was used to compare nested models.
(F) Survival of melanoma (pooled datasets 1, 2, 3, and 4) and bladder cancer (dataset 6) patients stratified in quantiles according to their COX-IS. Log-rank (Mantel-Cox) test.