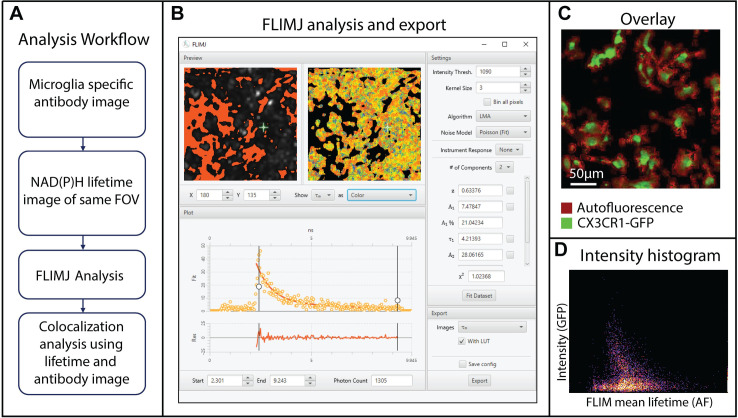
Fig 3. Microglia colocalization analysis using NADH FLIM and CX3CR1-GFP labels [39, 46].
A) The analysis workflow describing how microglia are visualized using a specific antibody, followed by NADH FLIM acquisition and FLIMJ analysis B) NADH FLIM data analysis using 2-component fit in FLIMJ-UI. Users can choose the intensity threshold, kernel size, fitting model, noise model, model restraints, and the number of components. The single curve fit is fast, and the “Fit Dataset” button performs fits for all the pixels. The fit result and fitting-parameters can be exported from the export tab on the lower right part of the UI. C) Overlaid images of antibody (green) and lifetime image (red) to show the pixels with overlapping NADH and GFP signal. D) Coloc2 analysis of mean lifetime and microglia antibody image.