Figure 1: The overall architecture.
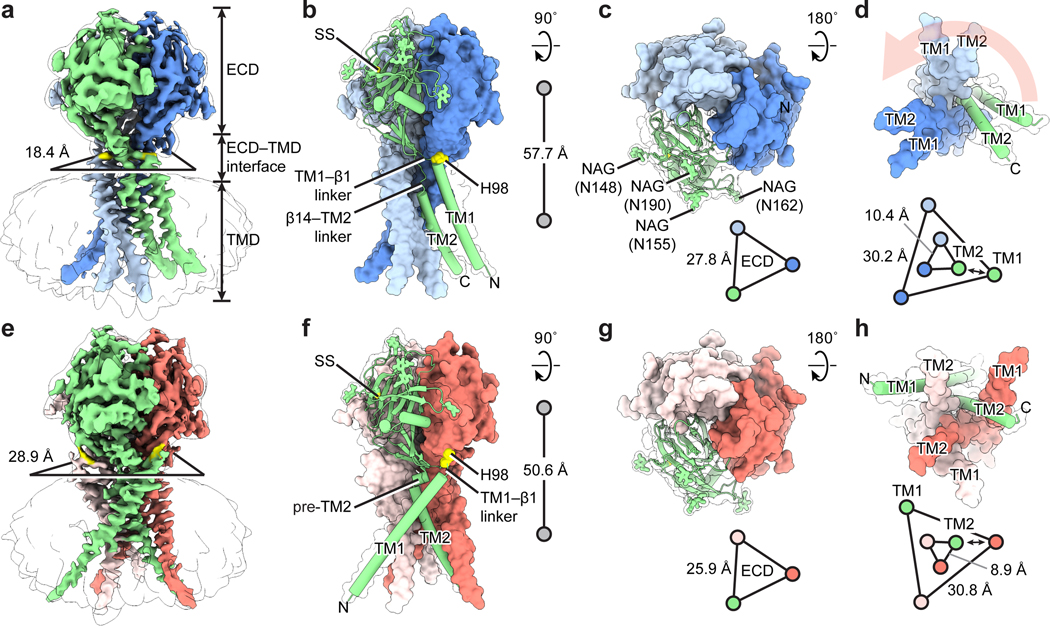
a and e, Cryo-EM maps of pH8–PAC and pH4–PAC viewed parallel to the membrane. The map refined without using a mask is shown as a transparent envelope. The horizontal dimension of the ECD–TMD interface is represented by the distance between the Cα atoms of adjacent H98. The density for H98 is colored in yellow in both maps. b and f, The atomic models for pH8–PAC and pH4–PAC. The green subunit is shown as a cartoon and the other two subunits are shown in surface representation. The center-of-mass distances between the ECD and the TMD are shown on the right. c and g, The TMDs of pH8–PAC and pH4–PAC viewed from the intracellular side. A light salmon arrow (panel c) indicates the rotation of TM1 of PAC after acidification to pH 4. The relative position and distance of TM1 and TM2, which are represented by the Cα atoms of I73 and K319, respectively, are shown at the bottom. The double-headed arrow indicates the interaction between TM1 and TM2. d and h, The ECDs of pH8–PAC and pH4–PAC viewed from the extracellular side. Four putative glycosylation sites (N148, N155, N162, and N190) are labeled in (d). The center-of-mass distance between the ECDs of each subunit is shown by the triangles.