Fig 6. Identification of optimal 8-mer and 9-mer peptide epitopes.
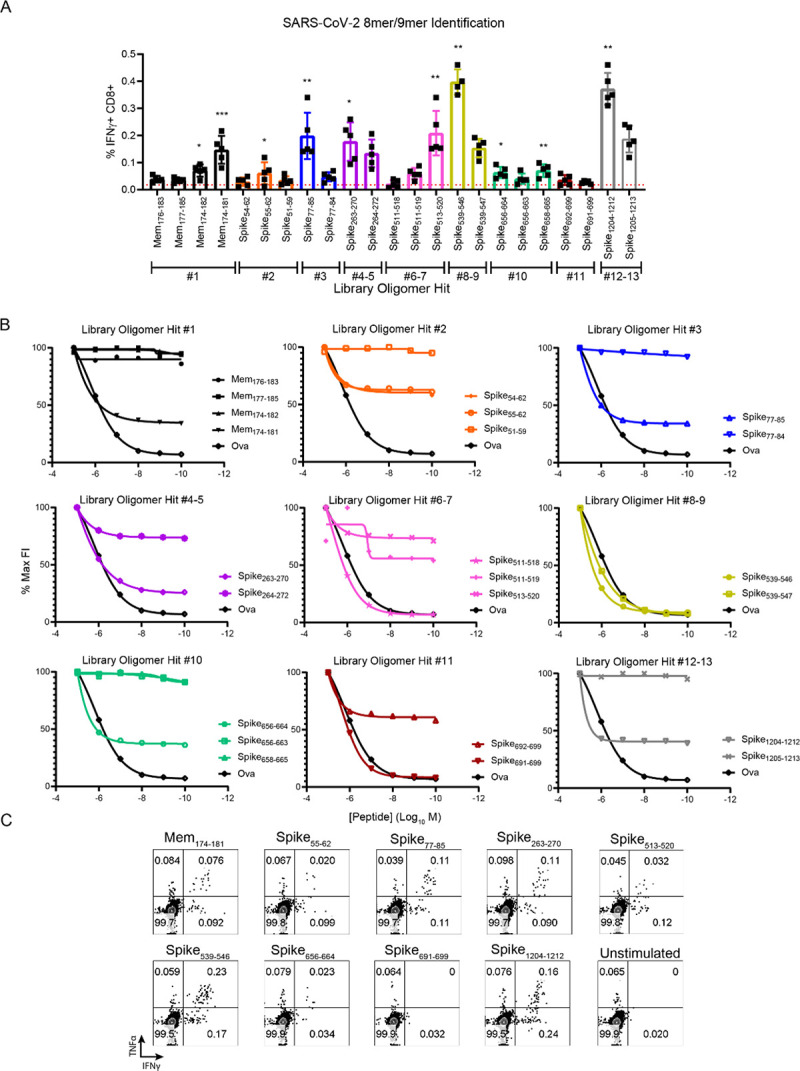
(A) T cell response to 8-mer or 9-mer peptides. Multiple SARS-CoV-2 8-mer or 9-mer peptide variants from each potential library hit were identified and purchased based on known Kb or Db anchor residues. Five days post boosted infection with SARS-CoV-2 following transfection with hACE2 mRNA, splenocytes were harvested and stimulated for 6 hours with each peptide in the presence of brefeldin A. After stimulation, cells were stained for flow cytometry to evaluate the frequency of responsive CD8+T cells by IFN-γ expression. Each color is indicative of a peptide variant being derived an individual potential library hit. (B) Kb RMA-S stabilization assay. To determine relative ability of individual peptide variants to stabilize the Kb molecule, decreasing concentrations of each peptide variant were incubated for 4 hours with TAP deficient RMA-S cells at 29 degrees C before being moved to 37 degrees C for 1 hour. Cells were then stained for either Kb or Db with fluorescently conjugated antibodies and geometric mean fluorescent intensity (gMFI) was measured using an Atttune focusing flow cytometer. Fluorescence index (FI) was determined by dividing the gMFI of cells pulsed with peptide by cells with no peptide. Data is presented as a percentage of the maximum FI for each peptide. As a positive control, the Kb restricted peptide Ovalbumin (SIINFEKL) was used. (C) Representative cytokine responses to each optimal epitope of mice transfected with hACE2 and infected with SARS-CoV-2 five days post boost.
