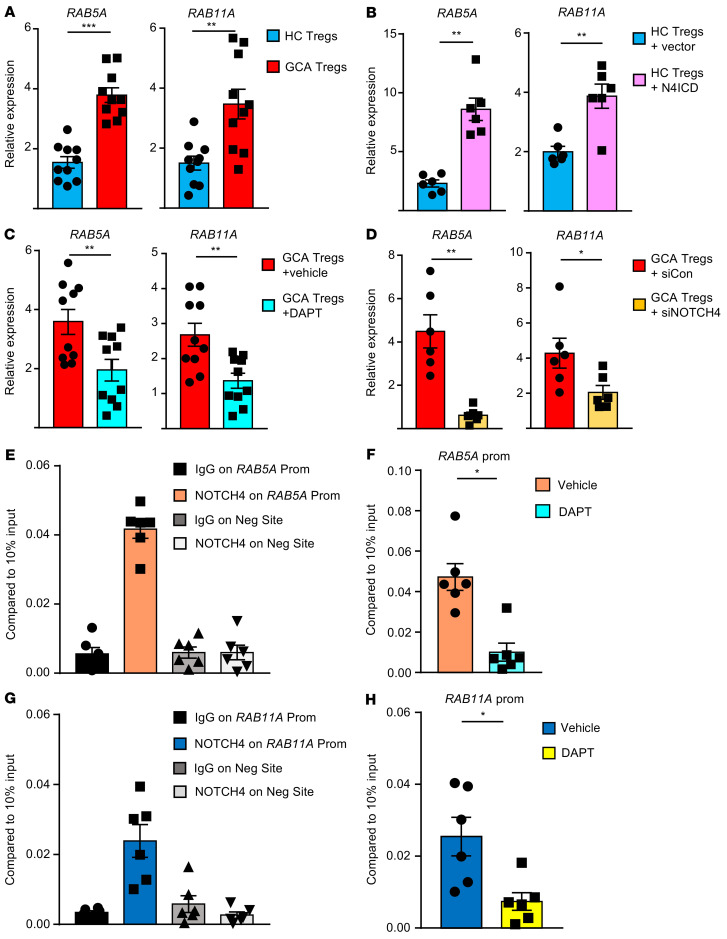
Figure 6. NOTCH4 signaling regulates RAB5A and RAB11A transcription.
(A) CD8+ Tregs were induced from controls or GCA patients. RAB5A and RAB11A transcripts were determined by q-PCR. Mean ± SEM from 10 samples each. (B) Healthy CD8+ Tregs were transfected with empty vector or a NOTCH4 ICD overexpression plasmid, and RAB5A and RAB11A transcript levels were determined by q-PCR. Mean ± SEM from 6 control and 6 GCA samples. (C) GCA CD8+ Tregs were treated with DAPT or vehicle. RAB5A and RAB11A mRNA were quantified by q-PCR. Mean ± SEM from 10 samples. (D) GCA CD8+ Tregs were transfected with control or NOTCH4 siRNA. RAB5A and RAB11A transcript expression was determined by q-PCR. Mean ± SEM from 6 samples. (E) CD8+ Tregs were generated from GCA patients. ChIP assays targeting NOTCH4 or control IgG were performed on the promoter of RAB5A or a negative site. The signal was normalized to 10% of input. Mean ± SEM from 6 samples. (F) GCA CD8+ Tregs were treated with DAPT or vehicle. ChIP assays targeting NOTCH4 were performed on the promoter of RAB5A. The signal was normalized to 10% of input. Mean ± SEM from 6 samples. (G) In GCA CD8+ Tregs, ChIP assays targeting NOTCH4 or control IgG were performed on the promoter of RAB11A or a negative site. The signal was normalized to 10% of input. Mean ± SEM from 6 samples. (H) CD8+ Tregs were treated with DAPT or vehicle. The occupancy of NOTCH4 on the RAB11A promoter was determined by ChIP assay and normalized to 10% of input. Mean ± SEM from 6 samples. CD8+ Treg cells were induced ex vivo for all experiments. *P < 0.05; **P < 0.01; ***P < 0.001 by unpaired Mann-Whitney-Wilcoxon rank test (A) or paired Mann-Whitney-Wilcoxon rank test (B–D, F, and H).