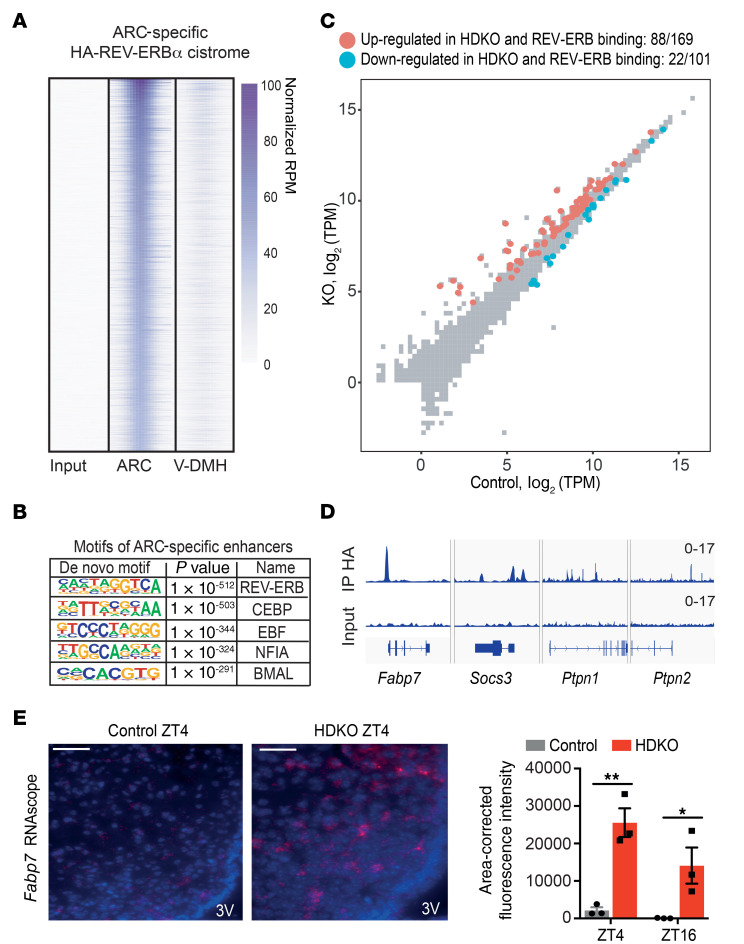
Figure 5. REV-ERBα directly regulates ARC transcriptome on HFD.
(A) HA-REV-ERBα cistrome specific to the ARC on HFD. ChIP-seq analysis of HA immunoprecipitation in the ARC from HA-REV-ERBα mice on HFD at ZT4 (reads per million mapped reads [RPM] > 1, >7-fold over input, FDR < 0.0001). Ten independent hypothalamic nuclei punches were pooled together (n = 1). (B) Motif analysis of ARC-specific cistrome of HA-REV-ERBα on HFD with default HOMER parameters. (C) Scatterplot analysis of the HFD-HDKO–specific ARC transcriptome at ZT4. The direct regulome was identified by the integration of the HA-REV-ERBα cistrome in the ARC on HFD (TPM > 0.1, FDR < 0.05 for differentially expressed genes; RPM > 1, >7-fold over input for peaks within 100 kb). (D) Genome browser view of HA-REV-ERBα ChIP-seq peaks in the ARC. (E) Fabp7 staining by RNAscope in the ARC from control and HDKO mice on HFD. Representative pictures at ZT4 and quantification at ZT4 and ZT16 are presented. Scale bars: 50 μm. 3V, third ventricle. Results were compared by 2-way ANOVA (interaction P = 0.1705) and Holm-Šidák multiple-comparison test. *P < 0.05, **P < 0.01.