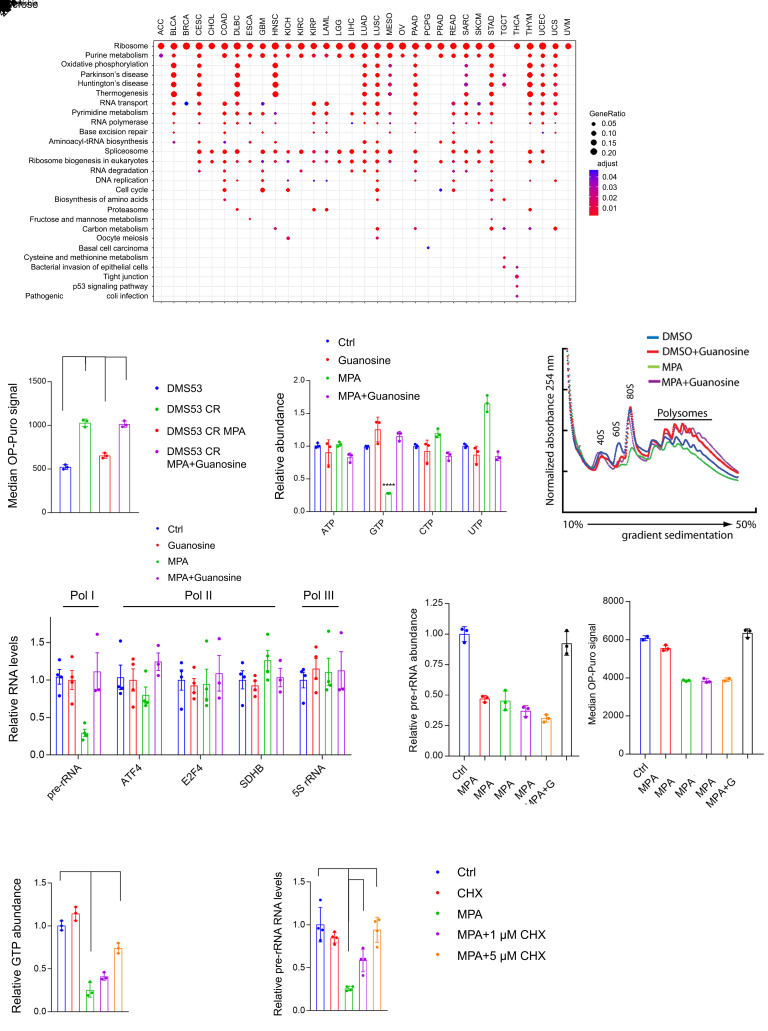
Figure 4. Enhanced de novo GTP synthesis promotes Pol I activity in chemoresistant SCLC cells.
(A) Gene sets associated with IMPDH2 mRNA across human cancers in the TCGA database. All KEGG gene sets were included in the analysis. (B) Median OP-Puro signal in DMS53 treatment-naive and chemoresistant cells treated with vehicle or 1 μM MPA, with or without 20 μM guanosine for 12 hours. ***P < 0.001. (C) ATP, GTP, CTP, and UTP levels in DMS53-CR treated with vehicle or 5 μM MPA, with or without 10 μM guanosine for 8 hours. **P < 0.01, ****P < 0.0001. (D) Ribosome abundance in DMS53-CR treated with vehicle, 1 μM MPA, with or without 20 μM guanosine for 48 hours. (E) Abundance of Pol I, II, or III transcripts in DMS53-CR cells treated with vehicle or 5 μM MPA with or without 20 μM guanosine for 8 hours. ***P < 0.001. (F) Abundance of pre-rRNA and median OP-Puro signal in H82 treated with vehicle or 1 μM MPA, with or without 10 μM guanosine. ****P < 0.0001. (G and H) Abundance of GTP and pre-rRNA in DMS53-CR treated with vehicle or 5 μM MPA, with or without 1–5 μM cycloheximide for 6 hours. **P < 0.01, ****P < 0.0001. Data are shown as mean and SD (B, C, and F–H), mean and SEM (E). Statistical significance was assessed using 1-way ANOVA with Tukey’s multiple-comparison test (B, C, and E–H). All experiments were repeated twice or more.