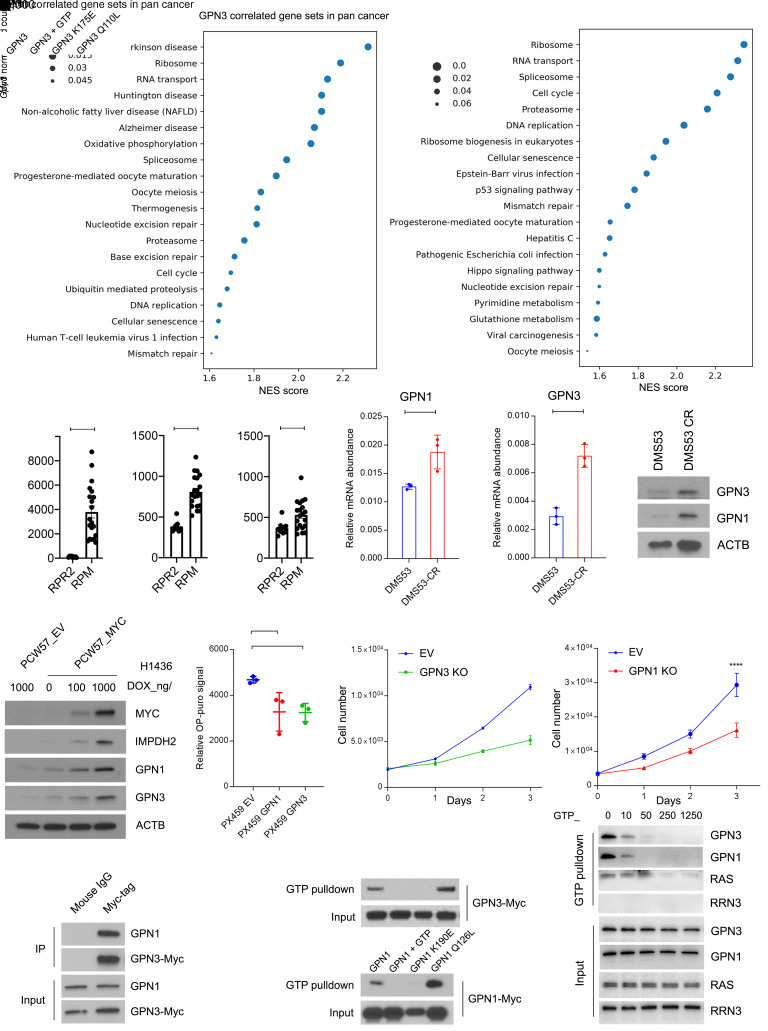
Figure 6. GTP abundance regulates Pol I function in Mychi cells in part through GPN1 and GPN3.
(A) Gene sets correlated with GPN3 or GPN1 mRNA from 9,879 tumors in the pan-cancer TCGA database. Gene set enrichment analysis was performed on the top 2% of genes positively correlated with GPN3 or GPN1. (B) mRNA abundance of Myc, Gpn1, and Gpn3 in SCLCs obtained from genetically engineered mouse models with mutant Trp53, Rb1, and Rbl2 (RPR2) and tumors with mutant Trp53 and Rb1 plus transgenic MycT58A (RPM). *P < 0.05, ****P < 0.0001. (C) GPN1 and GPN3 mRNA abundance in DMS53 and DMS53-CR. **P < 0.01, ***P < 0.001. (D) Abundance of GPN1 and GPN3 in DMS53 and DMS53-CR. (E) Abundance of MYC, IMPDH2, GPN1, and GPN3 in H1436 cells expressing dox-inducible empty vector (EV) or MYC treated with doxycycline at the indicated doses for 6 days. (F) OP-Puro signal in pooled H82 cells expressing an EV or with CRISPR/Cas9-mediated knockout of GPN3 or GPN1. **P < 0.01. (G) Proliferation of H82 pools shown in F. **P < 0.01, ****P < 0.0001. (H) Immunoprecipitation with anti-Myc-tag or mouse IgG followed by Western blot for GPN1 or Myc in H82 cells with CRISPR/Cas9-mediated GPN3 knockout and reexpression of wild-type GPN3. (I) GTP pulldown for wild-type or mutant isoforms of GPN3-Myc or GPN1-Myc. GTP was used to compete for binding of GPN1/3 to GTP-agarose beads. (J) GTP pulldown for GPN3, GPN1, RAS, or RRN3 using H82 lysates with increasing concentrations of GTP to compete for binding to GTP-agarose beads. Data are shown as mean and SD (B, C, F, and G). Statistical significance was assessed using a 2-tailed Student’s t test (B and C), 1-way ANOVA with Tukey’s multiple-comparison test (F and G). All experiments were repeated twice or more.