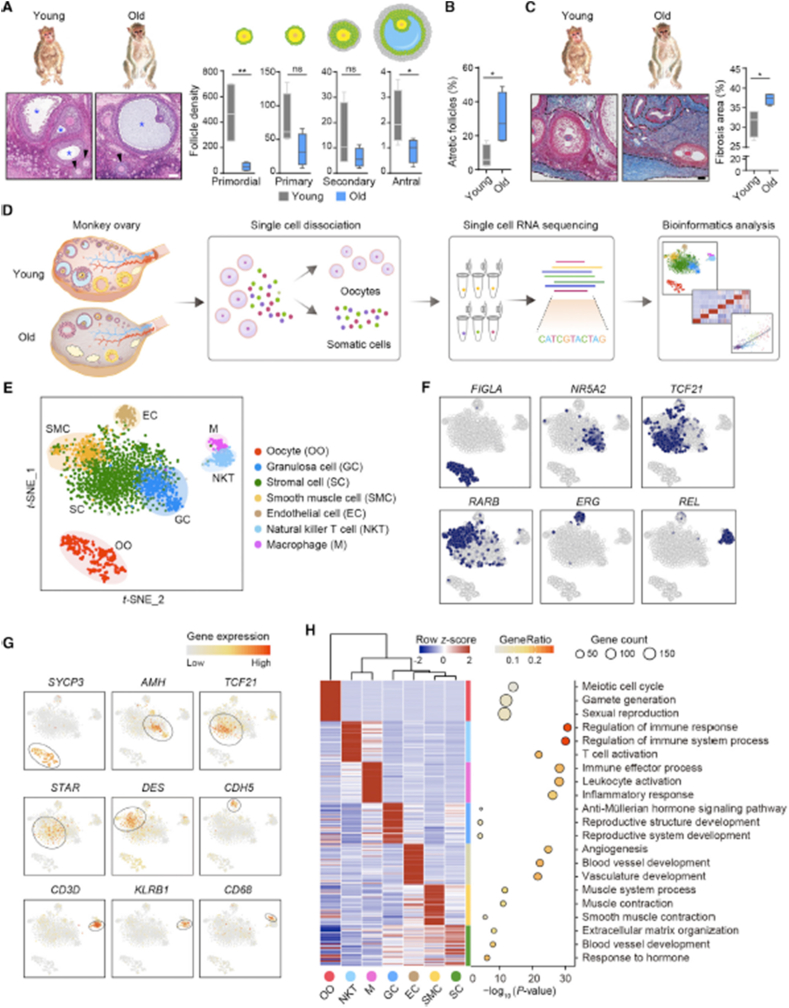
Fig. 4.
Distinct ovarian cell subpopulations with transcriptional signatures determined by single-cell RNA-Seq analysis. (A) Left: H&E stained sections of young and old monkey ovaries. Arrowheads and asterisks denote secondary and antral follicles, respectively. Right: density quantification of different stages of follicles indicated by the morphology shown by the cartoons. Scale bar, 100 mm n = 4 monkeys. ns, not significant, *p < 0.05, and **p < 0.01 (one-tailed t-test). (B) Percentage of atretic follicles with respect to total follicles based on H&E stained sections. n = 4 monkeys. *p < 0.05 (two-tailed t-test). (C) Masson's trichrome staining of young and old monkey ovaries. Dashed lines denote fibrotic areas. Scale bar, 100 mm n = 4 monkeys. *p < 0.05 (two-tailed t-test). (D)Flowchart overview of monkey ovarian scRNA-seq. (E) t-SNE plot showing seven ovarian cell types. (F) t-SNE plots characterizing representative transcriptional regulators for different cell types. Blue color denotes the cells with the activation of indicated transcriptional regulators. (G) t-SNE plots showing expression levels of oocyte and somatic cell marker genes. (H) Left: heatmap showing expression signatures of top 50 specifically expressed genes in each cell type; the value for each gene is a row-scaled Z score. Right: representative gene terms [76].