Figure 1.
Rtel1 Deletion Induces Transcriptional Changes That Are Independent of Its Role in Telomere Maintenance and That Overlap with Those Caused by G4 Stabilization
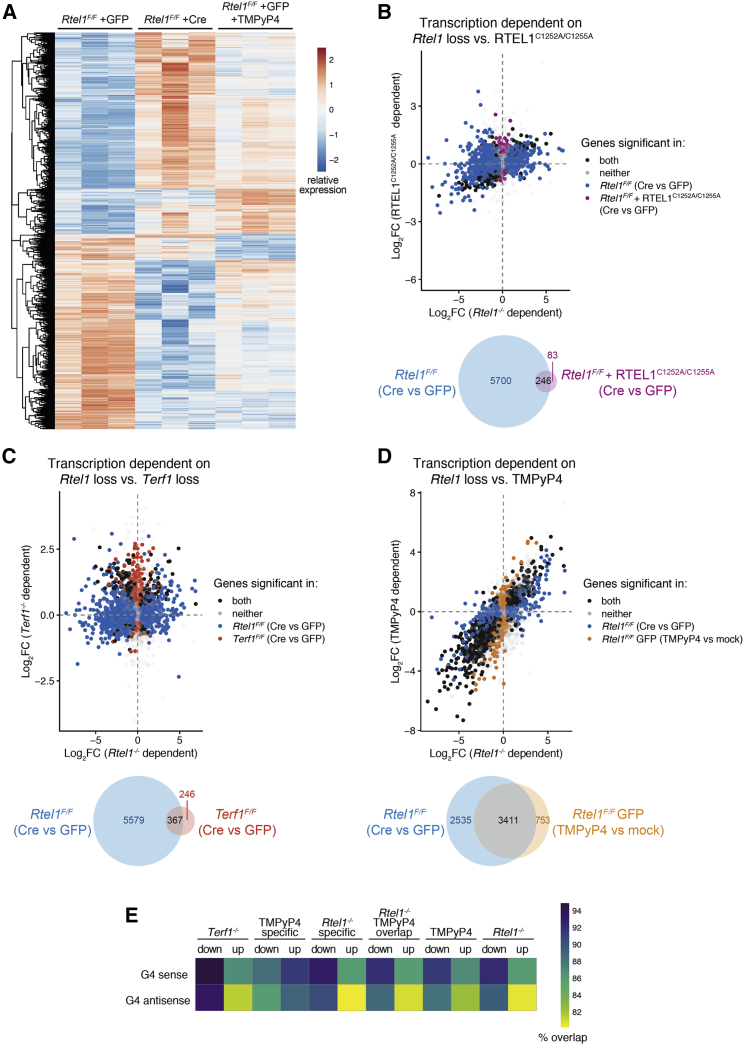
(A) RTEL1F/F MEFs were infected with GFP or Cre-GFP adenovirus and collected after 96 h or infected with GFP adenovirus for 48 h, treated with TMPyP4, and collected after 48 h. RNA was isolated, and gene expression levels were analyzed. Heatmap of norm transformed counts per significantly deregulated gene (p < 0.01 in any comparison between groups). Data are scaled by row and clustered with hierarchical clustering.
(B) Top: comparative differential gene expression between samples when Rtel1 was deleted (Rtel1F/F, Cre versus GFP) or contained a t-loop unwinding defect (Rtel1F/F + RTEL1C1252A/C1255A, Cre versus GFP). Differentially expressed genes are differentiated by their significance (p < 0.01) in the respective comparisons. Bottom: Venn diagram of differentially expressed genes.
(C) Top: comparative differential gene expression between samples when Rtel1 was deleted (Rtel1F/F, Cre versus GFP) or are shelterin defective (Terf1F/F, Cre versus GFP). Bottom: Venn diagram of differentially expressed genes.
(D) Top: comparative differential gene expression between samples when Rtel1 was deleted (Rtel1F/F, Cre versus GFP) or treated with TMPyP4 (Rtel1F/F GFP, TMPyP4 versus mock). Bottom: Venn diagram of differentially expressed genes.
(E) Heatmap of comparative proportions of G4-containing promoters in sense and antisense of differentially regulated genes following various treatments as indicated.