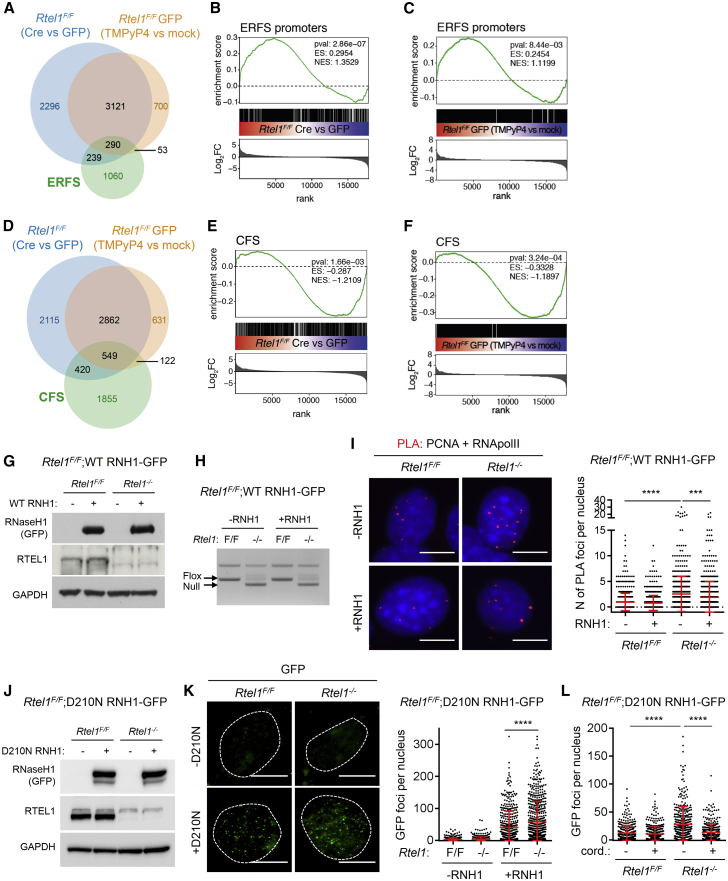
Figure 2.
RTEL1 Counteracts Transcription-Replication Conflicts by Regulating R-Loops
(A) Venn diagram of differentially regulated genes of samples with deleted Rtel1 (Rtel1F/F, Cre versus GFP) or samples treated with TMPyP4 (Rtel1F/F GFP, TMPyP4 versus mock), and their overlap with ERFSs.
(B) GSEA that shows enrichment of genes with ERFSs in their promoters. Genes are ranked dependent of logarithm of fold change (Log2FC) of differential expression with deleted Rtel1 (Rtel1F/F, Cre versus GFP), and the overall enrichment score (ES) and normalized enrichment score (NES) with the respective p value (pval) of the enrichment were determined.
(C) GSEA that shows enrichment of genes with ERFSs in their promoters. Genes are ranked dependent of Log2FC of differential expression upon TMPyP4 treatment, and the overall ES and NES with the respective pval of the enrichment were determined.
(D) Venn diagram of differentially regulated genes of samples with deleted Rtel1 (Rtel1F/F, Cre versus GFP) or treated with TMPyP4 (Rtel1F/F GFP, TMPyP4 versus mock), and their overlap with CFSs.
(E) GSEA that shows enrichment of genes overlapping with CFSs. Genes are ranked dependent of Log2FC of differential expression with deleted Rtel1 (Rtel1F/F, Cre versus GFP), and the overall ES and NES with the respective pval of the enrichment were determined.
(F) GSEA that shows enrichment of genes with CFSs in their promoters. Genes are ranked dependent of Log2FC of differential expression upon TMPyP4 treatment, and the overall ES and NES with the respective pval of the enrichment were determined.
(G) Rtel1F/F;WT RNH1-GFP MEFs were infected with GFP or Cre-GFP adenovirus. After 48 h, doxycycline was added, and cells were collected after 48 h. The cells were then lysed, and whole-cell extracts were analyzed by SDS-PAGE and immunoblotted for GFP, RTEL1, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
(H) Cells were treated as in (G), and genomic DNA was isolated and loss of Rtel1 was verified by PCR.
(I) Cells were treated as in (G), and the interaction between PCNA and RNA polymerase II (RNApolII) was assessed by PLA. Left: representative images of PLA. Right: quantification of PLA. Data are represented as mean ± SD (n = 4).
(J) Rtel1F/F;D210N RNH1-GFP MEFs were infected with red fluorescent protein (RFP) or improved Cre-RFP (iCre-RFP) adenovirus. After 48 h, doxycycline was added, and cells were collected after 48 h. The cells were then lysed, and whole-cell extracts were analyzed by SDS-PAGE and immunoblotted for GFP, RTEL1, and GAPDH.
(K) Cells were treated as in (J) and immunostained for GFP. Left: representative images of GFP immunofluorescence. Right: quantification of RNaseH1D210N-GFP foci per nucleus of cells. Data are represented as mean ± SD (n = 3).
(L) Rtel1F/F;D210N RNH1-GFP MEFs were infected with RFP or iCre-RFP adenovirus. After 96 h, cells were treated with cordycepin for 3.5 h and immunostained for GFP. Quantification of RNaseH1D210N-GFP foci per nucleus of cells. Data are represented as mean ± SD (n = 3).
The pvals were determined by unpaired t test, with ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001. Scale bars, 10 μm.