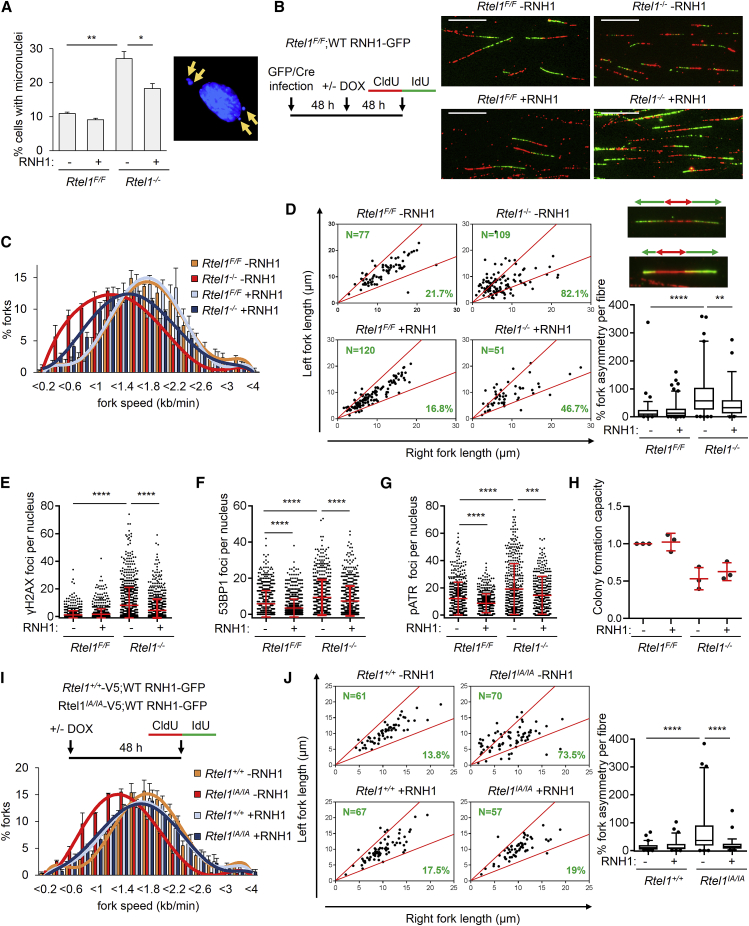
Figure 3.
Rtel1 Deletion Causes Genome-wide R-Loop-Dependent Replication Stress
(A) Rtel1F/F;WT RNH1-GFP MEFs were infected with GFP or Cre-GFP adenovirus. After 48 h, doxycycline was added, and cells were collected after 48 h. The cells were then fixed, and the percentage of cells with >1 micronucleus was quantified. Left: quantification. Right: representative images of micronuclei. Data are represented as mean ± SE (n = 3).
(B) Rtel1F/F;WT RNH1-GFP MEFs were treated as in (A), and a DNA fiber assay was performed. Left: experimental setup. Right: representative images of DNA fibers.
(C) Distribution of replication fork speeds of DNA fibers as prepared in (B). Data are represented as mean ± SE (n = 3).
(D) Left: scatterplot of fork asymmetry of DNA fibers as prepared in (B). Right, top: representative images of symmetric and asymmetric DNA fibers. Right, bottom: quantification of fork asymmetry of DNA fibers as prepared in (B) (n = 4).
(E) Rtel1F/F;WT RNH1-GFP MEFs were treated as in (A) and immunostained for γH2AX. Quantification of γH2AX foci per nucleus. Data are represented as mean ± SD (n = 3).
(F) Rtel1F/F;WT RNH1-GFP MEFs were treated as in (A) and immunostained for 53BP1. Quantification of 53BP1 foci per nucleus. Data are represented as mean ± SD (n = 3).
(G) Rtel1F/F;WT RNH1-GFP MEFs were treated as in (A) and immunostained for pATR-S428. Quantification of pATR-S428 foci per nucleus. Data are represented as mean ± SD (n = 3).
(H) Colony formation capacity in Rtel1F/F;WT RNH1-GFP MEFs infected with GFP or Cre-GFP adenovirus with or without doxycycline. Data are mean ± SD normalized to GFP and 0 μg/mL doxycycline condition (n = 3).
(I) Rtel1+/+-V5;WT RNH1-GFP and Rtel1IA/IA-V5;WT RNH1-GFP were treated with doxycycline for 48 h, and a DNA fiber assay was performed. Top: experimental setup. Bottom: distribution of replication fork speeds of DNA fibers. Data are represented as mean ± SE (n = 3).
(J) Left: scatterplot of fork asymmetry of DNA fibers prepared as in (I). Right, bottom: quantification of fork asymmetry of DNA fibers prepared as in (I) (n = 3).
In boxplots, horizontal line denotes the mean; whiskers denote the 5th and 95th percentiles. The pvals were determined by unpaired t test, with ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Scale bars, 10 μm.