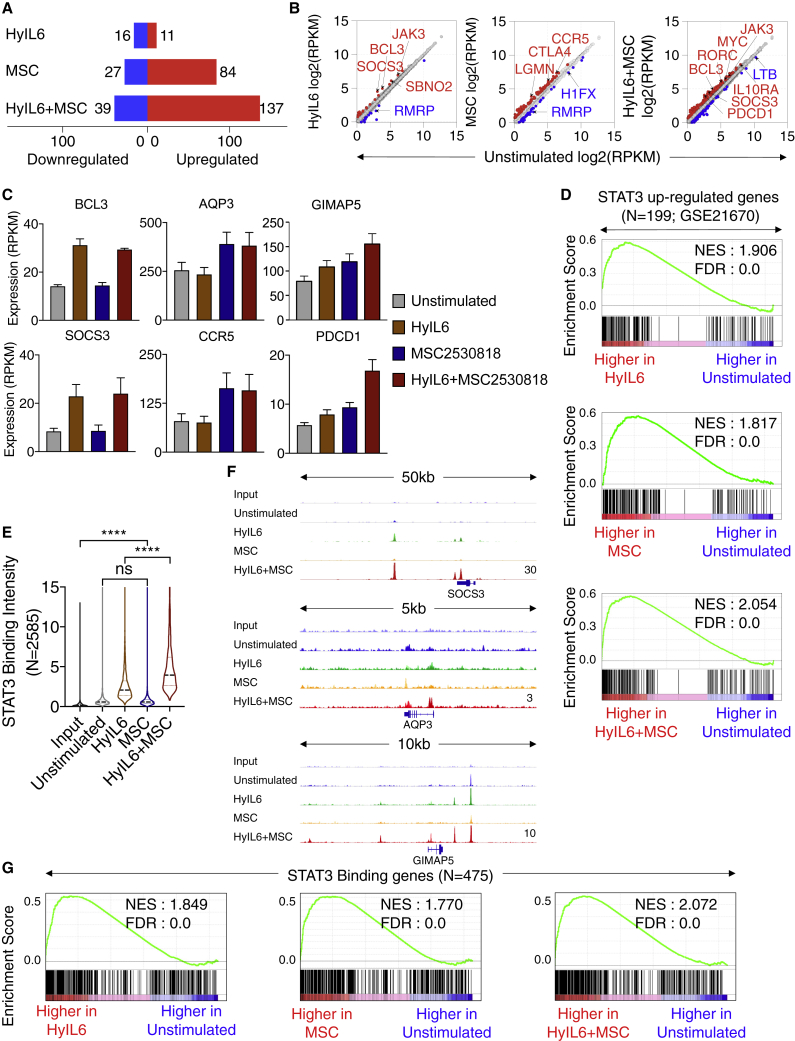
Figure 6.
Transcriptional Program Elicited by Interplay between HyIL-6 and CDK8 in Human Primary CD4+ Th-1 Cells
(A) Number of differentially expressed genes (DEGs; fold chang,e >1.5; p < 0.05) between unstimulated versus HyIL-6-, mesenchymal stem cell (MSC)-, or HyIL-6+MSC-stimulated Th-1 cells in three biological replicates.
(B) Scatterplot showing mean gene expression values (n = 3) before (x axis) and after indicated stimulation (y axis). Upregulated (red) and downregulated (blue) genes are highlighted.
(C) Representative gene expression across different stimulation. Bars show mean ± SEM.
(D) Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) plots for STAT3 upregulated genes (GEO: GSE21670) comparing stimulated versus unstimulated Th-1 transcriptomes. NES, normalized enrichment score; FDR, false discovery rate.
(E) Violin plot showing the mean STAT3 binding intensity in n = 2,585 STAT3-bound regions across different stimulations. Peaks are identified by comparing HyIL-6+MSC stimulation and input. The p values were determined by two-tailed Wilcoxon rank-sum test (∗∗∗∗p < 0.0001).
(F) Representative loci showing STAT3 binding across different stimulations. The height of the tracks are indicated at bottom-right corner of the plots.
(G) GSEA plots for 475 STAT3-bound genes comparing stimulated versus unstimulated Th-1 transcriptomes.