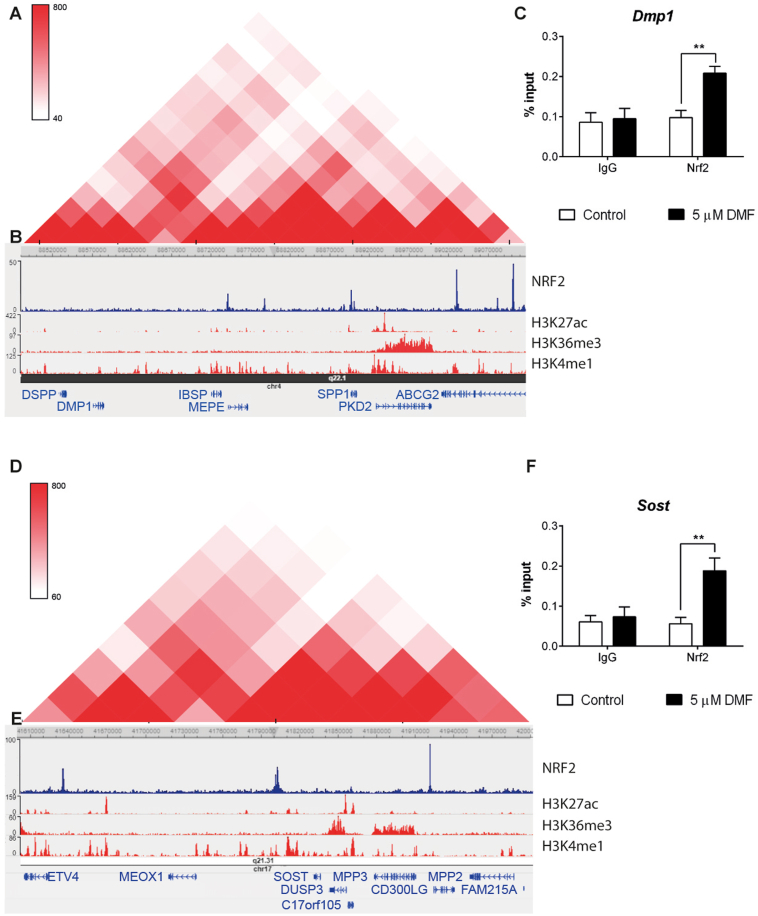
Fig. 6.
Osteocytic genes are organized in TADs regulated by NRF2 (A and D) Topological domain and looping structure indicated by Hi-C data from Dixon et al. (2015) in MSC. The schematic shows TAD (red-shaded triangle) containing the SIBLING family of proteins (A) and Sost (D). (B and E) ChIP-seq data showing NRF2 binding sites in A549 cells (data obtained from GSE113497), as well as active promoter/enhancer associated histone mark H3K27ac, gene body associated histone mark H3K36me3 and promoter associated histone mark H3K4me3 obtained from human osteoblast epigenome. (C and F). Chromatin immunoprecipitation from osteocytes cultured in the presence or absence of 5 μM DMF for 48 h. Results were normalized to input chromatin and plotted relative to untreated osteocytes (mean ± SEM; n = 5). *P < 0.05, **P < 0.01, and ***P < 0.001 using Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)