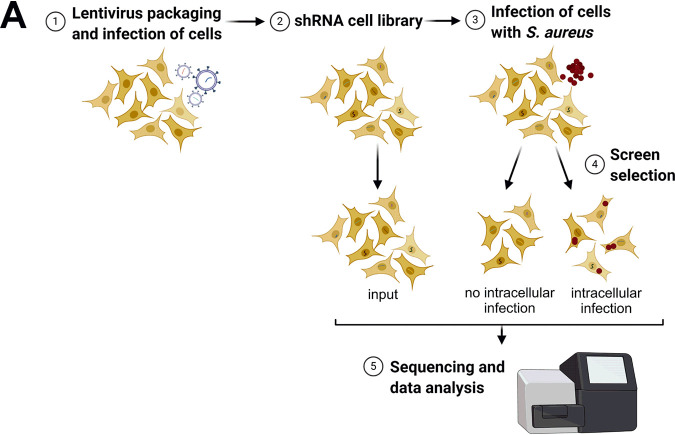
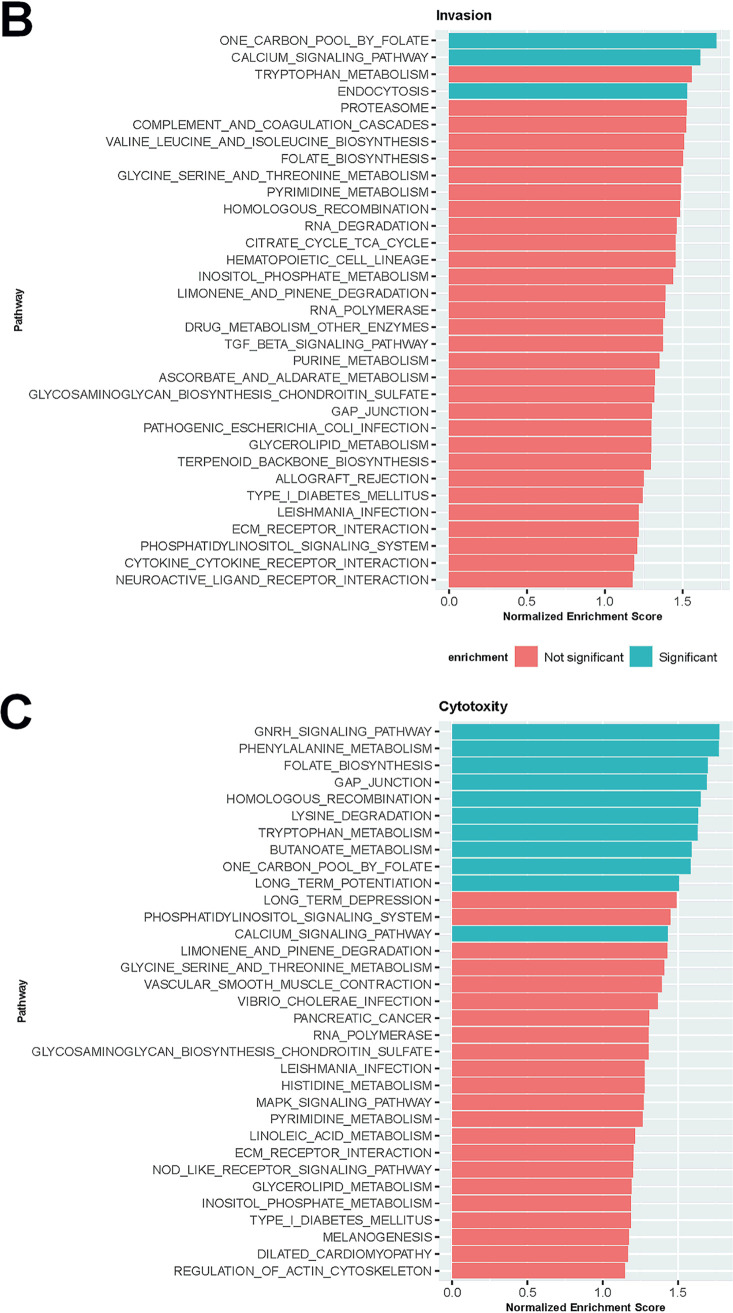
FIG 1.
Small hairpin RNA (shRNA) screen to identify host cell genes involved in S. aureus invasion and intracellular cytotoxicity. (A) Layout of shRNA screening approach. HeLa cells were first transduced with the shRNA library. Stable shRNA-HeLa cells were infected with S. aureus 6850 mRFP. At 4.5 hours postinfection (h p.i.), dead cells were removed, and both cells containing no intracellular bacteria and cells with intracellular S. aureus were sorted via fluorescence-activated cell sorting (FACS). Subsequently, genomic DNA was extracted and prepared for Illumina sequencing to determine enrichment or depletion of shRNAs in comparison to input HeLa cells, which were used as a negative control. (b, c) a detailed gene set enrichment analysis (GSEA) of genes identified by the shRNA screen to be implicated in S. aureus invasion (B) or cytotoxicity (C) revealed key pathways. Pathways with a P value lower than 0.02 are noted as significant.