Abstract
Metformin is associated with increased insulin sensitivity, whereas oral contraceptive pills (OCP) could increase the risk for type 2 diabetes (T2D) in women with polycystic ovary syndrome (PCOS). Certain miRNAs might serve as biomarkers for the risk of T2D. The aim of this study was to investigate changes in circulating miRNA levels during treatment with metformin and OCP in women with PCOS. Sixty-five women with PCOS according to Rotterdam criteria were randomized to metformin (2 g/day), metformin + OCP (150 mg desogestrel + 30 µg ethinylestradiol) or OCP alone for 12 months. Serum miRNA analysis was performed with individual RT-qPCR or Taqman low density array cards of 22 selected miRNAs previously related to PCOS, glucose and/or lipid metabolism. miR-122 and miR-29a levels were decreased after treatment with metformin compared with metformin + OCP and OCP group: miR-122: log2 difference −0.7 (P = 0.01) and −0.7 (P = 0.02), miR-29a: log2 difference −0.5 (P = 0.01) and −0.4 (P = 0.04), while miR-223 levels were decreased in the metformin + OCP group after treatment: log2 difference −0.5 (P = 0.02). During the treatment period, a significant weight loss was observed in the metformin group compared with the OCP group. In the OCP group, miRNA levels were unchanged during the treatment period. Levels of circulating miRNAs associated with lipid and glucose metabolism decreased during metformin treatment. Changes in miRNA levels in the metformin group could be explained by the simultaneous weight loss in the same group. These results support the notion that metformin treatment alone may be superior for metabolic health compared with OCP.
Key Words: metformin, polycystic ovary syndrome, microRNA, miR-122, biomarkers
Introduction
Polycystic ovary syndrome (PCOS) is characterized by hyperandrogenism, anovulation and polycystic ovarian morphology (1). It is well-established that PCOS has system-wide manifestations such as dyslipidemia, central obesity, hypertension and insulin resistance (1, 2), resulting in a three-fold increased risk of type 2 diabetes (T2D) (3). While obesity (4) and hyperandrogenism (2) as well as defects in the cellular insulin signaling pathway (5) play a central role, the primary cause of the association between insulin resistance and PCOS is still unclear.
Treatment of PCOS aims to decrease hyperandrogenism, regulate menstrual cycles and reduce risk of T2D by improving insulin sensitivity. First line treatment is lifestyle management (1), however medical treatment with metformin and/or oral contraceptive pills (OCPs) is often considered necessary. Metformin reduces insulin resistance, decreases insulin levels and improves dyslipidemia in women with PCOS, especially in women with BMI over 25 kg/m2 (6), while OCPs induce regular bleedings and increase steroid hormone binding globulin (SHBG) levels, which decreases levels of free testosterone and improves clinical hirsutism (7). OCP treatment is superior to metformin treatment regarding improved menstrual cycle irregularity, SHBG, free androgen index and total testosterone (6), while combination of metformin and OCP decreases the free androgen index, total testosterone and fasting glucose levels compared with metformin alone.
miRNAs are small non-coding RNAs, with a length of 19–22 nucleotides, suggested to have a physiological role in the development of diabetes (8, 9). Previous studies have shown an altered expression of miRNAs in insulin-sensitive tissues of obese patients (10, 11) and in patients with T2D (12, 13, 14). The role of miRNAs in metabolism and diabetes is quite complex (15) and involves many different tissues for example, liver, beta-cells, white and brown adipose tissue, as well as many different miRNAs. Some of these miRNAs (e.g. miR-122, miR-223) are believed to have a more pronounced role than others, as several studies confirm their associations (15, 16). miRNAs are produced in cells of various tissues and serve as intracellular regulators of gene expression, but are also present in blood and other body fluids serving as communicators between different insulin sensitive tissues (8). In blood, miRNAs are bound to proteins, lipoprotein complexes or contained in microvesicles (17), which protect miRNAs from degradation by ribonucleases. This property, along with resistance to freeze-thaw cycles (18), renders them suitable as biomarkers (17). Several studies have been dedicated to the detection of miRNAs as clinical biomarkers for PCOS (19, 20, 21, 22) or prognostic biomarkers for development of T2D (23, 24, 25). However, if miRNAs should serve as biomarkers for PCOS and T2D in a clinical setting, it is necessary to consider if standard treatment of PCOS could change the levels of circulating miRNAs during management of PCOS. Data on the effect of OCP treatment on the levels of circulating miRNAs in women is limited to a single study (20). A few clinical studies have addressed the impact of metformin on miRNA levels in patients with T2D (26, 27, 28, 29) but none have investigated the impact of metformin treatment on circulating miRNA levels in women with PCOS.
In this randomized study, we aimed to investigate the impact of metformin and OCP on the level of selected miRNAs in women with PCOS.
Methods and materials
Study design
This present study is based on analysis of serum from a biobank collected during a randomized controlled trial performed at Odense University Hospital, Denmark between 2007 and 2013 (registered at Clinical Trials.gov (NCT00451568)) (30). In this study, 90 women with PCOS were randomized to either metformin, OCP or metformin + OCP. The primary objective of the study (30) was to evaluate body composition during 12 months of treatment. A total of 65 women, 19 from the metformin group, 23 from the OCP group and 23 from the metformin + OCP group completed the study. All of the participants gave informed written consent to collection of biological material for future research. Permission to use the data has been granted by the Danish Data Protection Agency. The present study was approved by the Danish Scientific Ethical committee of Region Zealand (application nr. SJ-525).
The participants were all diagnosed with PCOS according to Rotterdam 2003 criteria (31). Patients with diabetes (fasting plasma glucose >7.0 mmol/L and/or glycated hemoglobin (HbA1c) >44 mmol/mol), elevated liver enzymes, renal dysfunction, congestive heart disease, depression, and eating disorders were not included in the study, neither were obese patients (BMI >35 kg/m2) nor patients with other contraindications for OCP. Patients were not included if they were pregnant or expressed a wish for conception during the study period. Patients paused OCP use for at least 3 months and metformin for at least 1 month before evaluation and no patients were treated with medicine known to affect hormonal or metabolic parameters. The participants were, subsequent to inclusion, randomized to 12 months treatment of metformin alone (1000 + 1000 mg/day) or OCP (150 mg desogestrel/130 g ethinylestradiol) or OCP + metformin.
During the trial, all participants received advice on lifestyle intervention. The participants attended examination at inclusion and at the end of trial as well as a visit for registration of side effects and compliance after 6 months. The clinical examination was performed on a random cycle day and included a DEXA scan, Ferriman-Gallwey (FG) score, WHR, height and weight. Fasting blood samples were analyzed for insulin, HbA1c, plasma glucose, electrolytes, and total and free testosterone. Homeostasis model assessment (HOMA) was calculated as fasting insulin · fasting blood glucose/22.5. Further details on assays and study protocol are given previously (30).
miRNA analysis with TaqMan custom cards
miRNA analysis was conducted on venous blood samples, collected in a procoagulant drying tube. Serum and cellular fractions were separated by centrifugation at 2000 g for 9 min. Serum was carefully removed leaving 0.5 mL in order to avoid disturbance of the interface. The serum was stored at −80°C until analysis.
Extraction of RNA: Total RNA was extracted from serum with TRI®Reagent LS (Sigma-Aldrich), according to manufacturers’ protocol. RNA concentration and purity was determined using and NanoDrop ND-1000 spectrophotometer (Thermo Scientific).
RT and qPCR: Total RNA was reverse transcribed using a 24 miRNA custom-designed TaqMan RT kit (ThermoFisher Scientific) according to the manufacturers’ protocol. miRNAs were subsequently analyzed using Human TaqMan low density array (TLDA) cards containing 24 different assays (ThermoFisher Scientific) according to manufacturers’ protocols. The TLDA card was designed to include a total of 21 miRNAs (40, 49, 62) and miRNAs reported in the literature to be associated with metabolic syndrome, insulin resistance or diabetes (Supplementary Table 1, see section on supplementary materials given at the end of this article). In addition to the 21 selected miRNAs, the card contained two spike in controls (ath-miR-159a and cel-miR-39) and one endogenous control (U6). The RT-product was pre-amplified with TaqMan® PreAmp Master Mix and miRNA PreAmp custom primer pools. Pre-amplified samples were diluted with 0.1 x TE buffer (pH 8.0) and stored at −80°C until analysis. All of the TLDA cards had same lot number. miR-122-5p was analyzed with individual RT-qPCR using specific RT- and qPCR primers, Multiscribe Reverse Transcriptase (ThermoFisher Scientific) and QuantiTect SYBR Green PCR master mix (Qiagen) according to manufacturers’ manual. Specific primer sequences are shown in Supplementary Table 2. PCR assays were performed in duplicates on a ViiA 7 real time PCR system and analyzed using Expression Suite software (v1.1, ThermoFisher Scientific, 2016).
All amplification plots were manually inspected and assays with absent or poor amplification or low fluorescent intensity were excluded for further analysis. Cycle threshold (Ct) >32 was considered undetectable following manufacturers’ guidelines and excluded from the analysis. Four (n = 4) samples at baseline (two from metformin group and two metformin + OCP group) and one (n = 1) sample (metformin + OCP group) at the end of study had low or no amplification and were not included in the analysis. Further one baseline sample was missing. miR-518f-3p showed only amplification in four samples at baseline and two at follow-up and the miRNA was therefore excluded from further analysis. U6 and miR-484 was determined as homogenous and stable controls with mean CT (s.d.) of 25.9 (1.9) and 20.3 (2.8), respectively, at baseline and 26.0 (1.7) and 19.8 (2.1), respectively, after intervention. The Ct-values were normalized to the geometric mean of ath-miR-159a (CT of 13.2 (0.53) and 13.2 (0.52) at baseline and after intervention, respectively), miR-484 and U6. Fold changes were expressed relative to baseline by the 2−ΔΔCt method (32).
Statistics
All data were analyzed using Statistical Packages for Social Sciences (SPSS, vers. 26, IBM) and GraphPad Prism (vers. 8.1.1, GraphPad Inc.). Correlation plots were generated with R (vers. 3.5.3), R Studio (vers. 1.2.1335) and statistical package Corrplot (vers. 0.84). All data were tested with the Shapiro–Wilk test for normal distribution. Non-normally distributed data were logarithmically transformed. Data are presented as medians and interquartile range (IQR) and miRNA levels as log fold change between indicated groups. For statistical analysis, grouped data were compared with Students’ t-test for unpaired and paired samples. For more than two comparisons, one-way ANOVA with Tukeys’ post hoc test was used. Spearmans’ correlation coefficient was used to assess correlations between miRNAs and relevant clinical variables. All correlations were subsequently adjusted for the effects of BMI and age. To assess the relationship between changes in miRNA and changes in clinical or biochemical parameters, the correlations between Δ-values of the five altered miRNAs and the Δ-values of the clinical measurements that changed significantly in any of the treatment groups during the study (Δ-weight, Δ-BMI, Δ-fat arms, Δ-fat trunk, Δ-fat legs, Δ-free testosterone, Δ-SHBG, Δ-insulin, Δ-C-peptide, Δ-FG-score), was used. The Δ-values were calculated as the post-treatment level minus the pre-treatment level of each analyzed value. Missing data were handled with pairwise deletion. The Bonferroni correction is used to adjust for multiple testing and is added after each P-value as (padjusted). An adjusted P-value of <0.05 was considered statistically significant.
Results
BMI, age, clinical and biochemical measurements were comparable at baseline in the three treatment groups (Table 1). At the end of the 12 months trial, metformin and metformin + OCP treatments were superior to OCP alone regarding weight loss and decreased regional fat free mass, whereas OCP treatment significantly increased SHBG levels and decreased free testosterone. Combined treatment with metformin and OCP improved body composition and FG-score compared with OCP, and SHBG-levels increased by OCP or metformin + OCP treatment compared with metformin alone (30). The decrease in BMI and weight in the metformin and metformin + OCP group was equally distributed during the 12 months (30).
Table 1.
Baseline characteristics of the participants.
| Metformin (n = 19) | Metformin and OCP (n = 23) | OCP (n = 23) | |
|---|---|---|---|
| Age (years) | 31 (24–33) | 30 (24–31) | 28 (25–32) |
| Weight (kg) | 73.6 (69.2–83.5) | 80.2 (70.5–86.0) | 86.0 (62.1–88.8) |
| BMI (kg/m2) | 26.0 (24.1–29.6) | 27.6 (24.3–31.3) | 28.0 (22.9–31.8) |
| Waist-hip ratio | 0.84 (0.76–0.88) | 0.84 (0.77–0.88) | 0.86 (0.76–0.87) |
| Total testosterone (nmol/L) | 2.0 (1.3–2.9) | 1.6 (1.3– 2.3) | 1.7 (1.4–2.7) |
| Free testosterone (nmol/L) | 0.035 (0.027–0.050) | 0.029 (0.022–0.045) | 0.033 (0.019–0.053) |
| Ferriman Gallway-score | 5 (0–11) | 8 (3–12) | 5 (2–9) |
| SHBG (nmol/L) | 44.0 (32.0–62.0) | 47.0 (32.0–72.0) | 52.0 (36.0–82.0) |
| Glucose (mmol/L) | 5.3 (4.9–5.4) | 5.2 (5.0–5.6) | 5.2 (5.0–5.6) |
| Insulin (pmol/L) | 61 (49–91) | 70 (44–79) | 67 (50–120) |
| C-peptide (pmol/L) | 687 (618–991) | 673 (589–787) | 748 (582–972) |
| HOMA-IR | 14.3 (11.1–19.3) | 15.3 (11.2–19.5) | 16.0 (12.1–28.9) |
| Total cholesterol (mmol/L) | 4.7 (4.4–5.1) | 4.4 (4.0 –4.7) | 4.3 (3.9–5.1) |
| LDL cholesterol (mmol/L) | 2.8 (2.6–3.3) | 2.6 (2.4–2.8) | 2.6 (2.2–3.3) |
| HDL cholesterol (mmol/L) | 1.5 (1.2–1.8) | 1.3 (1.2–1.5) | 1.3 (1.2–1.4) |
| Triglyceride (mmol/L) | 1.0 (0.7–1.3) | 0.9 (0.7–1.3) | 0.9 (0.6–1.4) |
| Fat, arms (kg) | 3.2 (2.5–4.0) | 2.8 (2.3–3.8) | 3.4 (2.5–4.3) |
| Fat, legs (kg) | 10.5 (8.2–12.6) | 9.6 (8.6–11.3) | 11.2 ( 7.0–14.0) |
| Fat, trunk (kg) | 15.9 (13.4–19.5) | 16.4 (11.0–19.6) | 18.9 (12.1–22.0) |
Data are presented as medians (IQR). There were no differences between the groups in any of the variables (one-way ANOVA, Tukey post hoc).
HDL, high density lipoprotein; HOMA-IR, homeostatic model assessment for insulin resistance; LDL, low density lipoprotein; SHBG, sex hormone binding globulin.
Changes in circulating miRNA levels after intervention
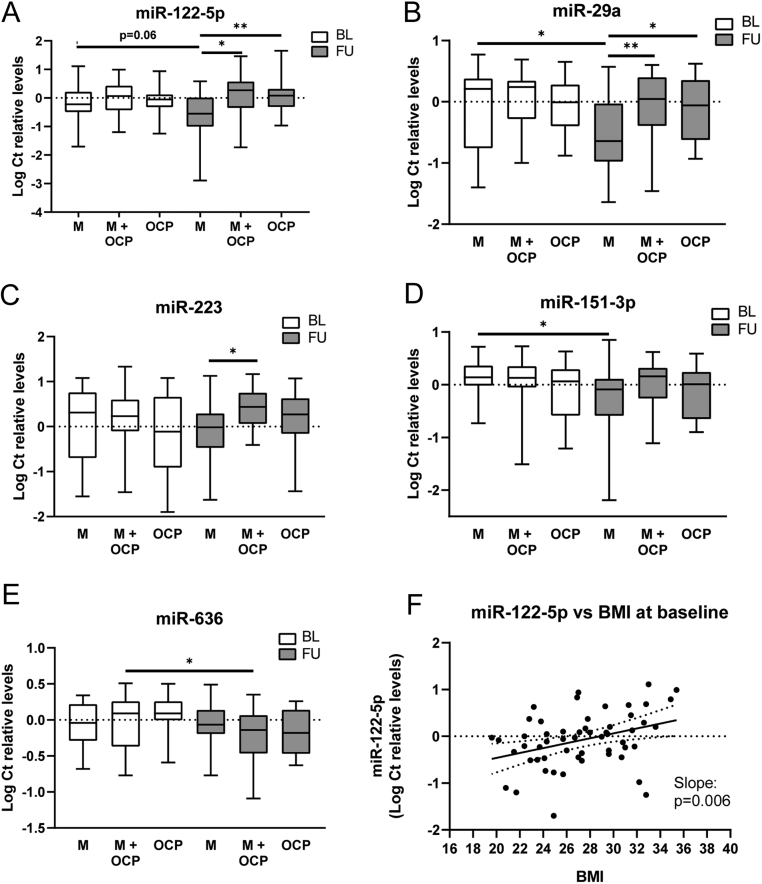
At baseline, circulating levels of all the assayed miRNAs were similar between the groups (metformin, metformin + OCP and OCP) (Supplementary Table 2). At the end of the trial, three miRNAs, miR-122, miR-29a and miR-223, were significantly decreased in the metformin treated group compared with the OCP treated group and the metformin + OCP group (Fig. 1 and Supplementary Table 3A). Circulating miR-122 levels were significantly decreased in the metformin treated patients compared with both metformin + OCP and the OCP treated patients (P = 0.01 (Padjusted = 0.22) and P = 0.02 (Padjusted = 0.44), respectively) (Fig. 1A). miR-29a levels were similarly decreased in the metformin group compared with both metformin + OCP (P = 0.01 (Padjusted = 0.22)) and OCP treatment (P = 0.04 (Padjusted = 0.88)) (Fig. 1B). miR-223 levels were significantly decreased in the metformin group compared with the metformin + OCP-group, while the OCP group had intermediate levels of miR-223 (P = 0.02 (Padjusted = 0.44)) (Fig. 1C).
Figure 1.
Panel A–E displays boxplot of log Ct relative levels of the five miRNAs that were significantly altered at the end of the study. White boxes are baseline levels and grey boxes are levels at the end of the study in the respective treatment groups: M, Metformin; M + OCP, Metformin + Oral Contraceptives and OCP, Oral Contraceptives. Differences are determined with one-way-ANOVA and Tukey’s post hoc test or paired sample t-test with adjustment for multiple groups. Panel F: Scatterplot of miR-122-5p levels plotted against BMI for all participants at baseline with regression line and CI. *P < 0.05, **P < 0.01.
Using paired sample testing to compare miRNA levels within each group of patients, at baseline with levels at the end of trial, we found that miR-29a and two additional miRNAs were decreased: miR-29a was decreased in the metformin group (mean log change relative to baseline = −0.50, s.e.m. = 0.21, P = 0.03) and a similar decrease was observed for miR-151-3p in the metformin treated group (mean log change relative to baseline = −0.47, s.e.m. = 0.19, P = 0.03, respectively) (Fig. 1B, D and Supplementary Table 3B). The change in miR-122 during the study was near significant in the metformin group (mean log difference = −0.47, s.e.m. = 0.24, P = 0.06) (Fig. 1A and Supplementary Table 3B). In the metformin + OCP group, only miR-636 decreased significantly (mean log change relative to baseline = −0.23, s.e.m. = 0.08, P = 0.01) at the end of the study compared to baseline (Fig. 1E and Supplementary Table 3B). miR-223 did not change significantly in any of the groups from baseline to follow-up. OCP treatment alone did not change levels of any of the selected miRNAs during the study.
miRNAs were correlated with BMI and indices of body fat distribution
Of the five altered miRNAs (miR-122, miR-151-5p, miR-223, miR-29a and miR-636), only levels of miR-122 and miR-223 were correlated with clinical or biochemical measurements at baseline (Supplementary Table 4). However, when adjusting for age and BMI these became non-significant. miR-122-5p was significantly correlated with BMI after adjustment for age (Spearman’s rho = 0.36, P = 0.006) at baseline. A scatterplot of BMI and miR-122 with regression line is shown in Fig. 1F. Both miR-122 and miR-223 were significantly upregulated in participants with BMI >25 kg/m2 at baseline (P = 0.01 and P = 0.03, respectively), while miR-29a levels were not significantly different in overweight subjects compared with normal weight subjects.
To assess the association between changes in miRNAs and changes in clinical or biochemical parameters, correlation analysis with Δ-values for the five altered miRNAs and Δ-values of clinical and biochemical measurements was used (Table 2). In these analysis, only the correlation between ΔFG-score was significantly associated with ΔmiR-29a (rho: −0.277 P = 0.04 after adjustment, respectively). The analysis did not reveal any further significant correlations, when adjusted for BMI at inclusion or adjusted for treatment group (Table 2).
Table 2.
Correlations between Δ-values.
| ΔmiR-122-5p | ΔmiR-151-3p | ΔmiR-223 | ΔmiR-29a | ΔmiR-636 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rho | P | Rho | P | Rho | P | Rho | P | Rho | P | |
| Δ weight | 0.14 | 0.27 | 0.10 | 0.47 | −0.01 | 0.99 | 0.01 | 0.96 | −0.21 | 0.12 |
| Δ BMI | 0.13 | 0.29 | 0.10 | 0.47 | −0.02 | 0.88 | −0.01 | 0.96 | −0.21 | 0.12 |
| Δ free T | −0.23 | 0.07 | −0.14 | 0.29 | −0.13 | 0.32 | −0.25 | 0.05 | 0.05 | 0.74 |
| Δ SHBG | 0.13 | 0.33 | 0.07 | 0.63 | 0.07 | 0.58 | 0.11 | 0.40 | −0.11 | 0.44 |
| Δ FG-score | −0.30* | 0.02 | −0.18 | 0.20 | −0.23 | 0.07 | −0.35** | 0.01 | 0.17 | 0.23 |
| Δ fat, arms | −0.01 | 0.98 | −0.06 | 0.65 | −0.04 | 0.78 | −0.13 | 0.34 | −0.22 | 0.11 |
| Δ fat, legs | 0.19 | 0.13 | 0.06 | 0.69 | 0.01 | 0.99 | 0.03 | 0.83 | −0.28* | 0.04 |
| Δ fat, trunk | 0.13 | 0.32 | 0.03 | 0.80 | 0.07 | 0.62 | −0.01 | 0.96 | −0.29* | 0.03 |
| Δ insulin | 0.18 | 0.17 | 0.18 | 0.20 | −0.07 | 0.61 | −0.03 | 0.82 | −0.14 | 0.33 |
| Δ c-peptide | 0.17 | 0.20 | 0.20 | 0.14 | −0.02 | 0.87 | −0.03 | 0.84 | −0.08 | 0.58 |
Correlations between Δ values for significantly altered miRNAs and Δ values for significantly altered clinical measurements during the study assessed with Spearman’s Rho. Unadjusted. The correlations between Δ miR-636 and Δ fat legs and Δ fat trunk, respectively, were insignificant when adjusting for BMI at inclusion and treatment group. Similar, was the correlation between Δ miR-122 and Δ FG score insignificant after adjustment, while the correlation between Δ miR-29a and Δ FG score remained significant.
*P < 0.05, **P < 0.01.
FG, Ferriman Gallwey; SHBG, sex hormone binding globulin; T, testosterone.
The levels of four of the five miRNAs, whose circulating levels were altered during the study, were highly positively correlated (Fig. 2). The remaining miRNAs tested also showed close miRNA:miRNA correlations. Supplementary Fig. 1 displays the miRNA correlation pattern showing three clusters of inter-correlated miRNAs, with only few miRNAs not having their circulating levels correlated with other miRNAs.
Figure 2.
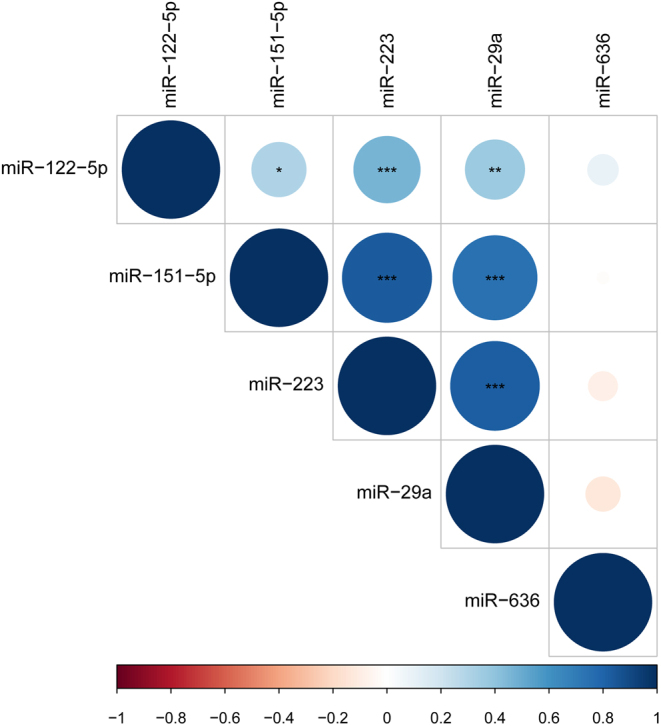
Plot of miRNA:miRNA correlations of the five altered miRNAs at baseline. The intensity of the color and size of the circles represent the Spearman rho-value. Blue colors are positive correlations, red colors are negative correlations. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
In this randomized controlled study, we investigated the effect of 12 months of treatment with metformin and OCP alone or in combination on circulating levels of 22 miRNAs, selected from the literature and from an in house pilot study of PCOS women. Metformin treatment was associated with decreased levels of three miRNAs, miR-122, miR-29a and miR-223 compared with OCP or metformin + OCP. In addition, one miRNA, miR-151-3p changed within the metformin group only and another, miR-636, within the metformin + OCP only from baseline to the end of study. None of the miRNAs were affected by OCP treatment alone. We are not aware of any previous study that has investigated the effect of metformin, OCP or metformin + OCP treatment on circulating miRNA levels in women with PCOS.
Our finding of decreased levels of miR-122 during metformin treatment is in agreement with previous studies of miRNAs in T2D, obesity and the metabolic syndrome: miR-122 was early described as a liver-specific miRNA in both mice and humans. Further it is an important regulator of cholesterol and fatty acid metabolism, as inhibition of miR-122 in both normal and high fat diet fed mice decreased plasma cholesterol levels and increased hepatic fatty acid oxidation (33, 34, 35, 36). Other studies have confirmed the role of miR-122 in clinical settings (23, 37), as miR-122 is decreased after gastric bypass-surgery (38) and after weight loss (39). The correlations between miR-122 and serum cholesterols were not strong in our study and not significant when adjusting for BMI and age. This could be due to our cohort being relatively young and lean compared to other cohorts in previous studies (23, 37). In our study we found miR-122 significantly correlated with BMI at baseline and upregulated in patients with BMI over 25. This is in accordance with previous clinical studies in which miR-122 has been suggested as a marker of metabolic health, as miR-122 was upregulated in patients with T2D (23) and obesity (37) compared with controls, and circulating miR-122 was decreased following a controlled weight loss (40). In agreement with this hypothesis, we found that miR-122 was significantly decreased in the metformin group at follow-up. The decrease in miR-122 levels may be due to weight loss, however, the changes in miR-122 and ΔBMI were not significantly associated, which could be due to small group sizes.
The mechanism for decreased miR-122 during metformin treatment are to date not clear. Studies of molecular mechanisms during metformin treatment have shown that metformin increases the levels of the miRNA processing protein DICER1 both in mice and humans with diabetes (41). Metformin treatment causes a disruption of the interaction between the RNA binding protein, AU-rich element binding factor 1 (AUF1), and DICER1 mRNA. This stabilizes DICER1 mRNA, allows translation and thereby causing a more general upregulation of all miRNAs. The same study noted a significant upregulation of miR-122 in liver tissue of mice treated with metformin compared with no treatment, but not compared with diet restriction (41). This contradicts our findings, but intracellular regulation of the levels of a specific miRNA could be different from the release of miRNA to the circulation, as the pool of circulating miRNAs both consists of actively secreted miRNAs and leakage from broken cells. One other study have described that extracellular vesicles derived from plasma of T2D patients treated with metformin contain lower levels of miR-122 than matched T2D patients not treated with metformin (27).
In this present study, miR-29a was decreased after metformin treatment compared with OCPs. miR-29a is reported as an important regulator of glucose metabolism in various tissues (42, 43, 44), and circulating miR-29a levels are increased in patients with T2D (45). It has been suggested that increased levels of miR-29a is promoting T2D by decreasing insulin secretion from beta cells (46, 47) and that miR-29 induces insulin resistance in muscle and adipose tissues (42). The mechanism for insulin resistance was both indirectly by targeting peroxisome proliferator-activated receptor delta (PPARD) mRNA, and directly by targeting solute carrier family 2 member 4 (SLC2A4) mRNA – the gene encoding GLUT4, the main insulin-mediated glucose transporter in adipose tissue (42). Further, miR-29a has also been demonstrated to have a key regulatory role in lipid metabolism in liver (48). How metformin treatment affects miR-29a is still unknown and no in vivo study has to date described miR-29a during metformin treatment. However, one rat study showed that increased circulating miR-29a was normalized in diabetic fatty rats after treatment with pioglitazone – another insulin sensitizing agent (48). miR-29a has also previously been associated with PCOS: In a study of follicle fluid from 49 women with PCOS miR-29a was significantly decreased both in hyperandrogenic and normoandrogenic women with PCOS (49).
In this study (49), women with PCOS also presented with lover levels of miR-151-3p. We found a decrease from baseline to end of study, of miR-151-3p in the metformin group, while miR-636 decreased in the metformin + OCP group. miR-151-3p has also, previously, been associated with diabetes, as decreased levels were found in late stage diabetes in Zucker diabetic fatty rats (50). In vivo studies of miR-636 are few, but one study demonstrated an association between miR-636 and restoration of liver damage, and that miR-636 are decreasing during antiviral treatment of hepatitis C virus (51) and two other studies have associated miR-636 with diabetic nephropathy (52, 53).
We found a decreased level of miR-223 in the metformin group. miR-223 is, similar to miR-122 and miR29a, also expressed in the liver and involved in cholesterol metabolism, by inhibiting HDL-cholesterol uptake in hepatic cells (54). The association between insulin resistance and miR-223 is also well-established; miR-223 is upregulated in adipose tissue in women with insulin resistance, both with and without PCOS (55), but miR-223 is upregulated also in serum from patients with diabetes and obesity (56, 57). miR-223 was suggested to target GLUT4 (58) and upregulation of miR-223 was shown to decrease GLUT4 protein content and glucose uptake in adipose tissue (55). These results were in accordance with our results, as we found a decreased level of miR-223 in the metformin group. Women in the metformin group obtained a significant weight loss and decreased regional fat mass, and thereby possibly improved their metabolic health. We did not observe a significant change in fasting levels of neither insulin nor C-peptide during the study in the metformin group, however both of these were significantly decreased in the metformin + OCP group (30). Thus, miR-122, miR-29a and miR-223 increase with obesity and/or hyperglycemia and their intracellular actions are involved in development of insulin resistance. Although miR-223 did not significantly decrease over time in the metformin treated PCOS women, miR-223 was lower in the metformin treated PCOS women than in the metformin + OCP treated women after the 12-month follow-up.
We observed close miRNA:miRNA correlations among the miRNAs, which changed during treatment, but all of the analyzed miRNAs correlate with each other in several large clusters (Supplementary Fig. 1). The origin of these clusters is uncertain, but could reflect miRNAs secreted from different tissues (59). However, the correlations between circulating miRNA levels and clinical or biochemical measures, although significant, were not strong, but this could be due to our limited sample size. The limited sample size is caused by the drop-out from each group (metformin = 11, metformin + OCP = 7, OCP = 7). Drop-rates were however, not statistically different between groups (P = 0.4), but should constitute a general limitation of the study. Regarding the study design, a control group of no treatment would have been suitable for miRNA analysis, but as this study is a post hoc study, no such exists.
Finally as a limitation, when applying the Bonferroni correction for multiple testing, none of the differences between the groups are significant and readers should therefore interpret the results with this in mind.
To our knowledge, this is the first study of miRNA during treatment with both metformin and OCP in women with PCOS. There are several studies on circulating miRNAs during metformin treatment in different cancers (60) and four studies exist of circulating serum or plasma in patients with T2D (26, 27, 28, 29). However, these studies have different methodological approaches and report changes in a variety of miRNA species (Supplementary Table 1). Only one previous study has investigated the effect of anti-androgen treatment (ethinyl-estradiol and cyproterone acetate) on serum miRNAs levels in a small group of seven women with PCOS (20). This study found that four months of anti-androgen treatment increased levels of miR-155 and that miR-155 was negatively correlated with androstenedione and 17OH-progesterone. Unfortunately, we have not tested miR-155 in our study, as the custom TaqMan array cards for our study was designed prior to this publication.
In conclusion, we investigated the effect of different treatment modalities of PCOS on miRNA levels. We did not see an effect of OCP-treatment on miRNA levels. These results suggest that while PCOS is a condition of hyperandrogenism, pharmacological intervention with OCPs to decrease hyperandrogenism did not change miRNAs previously associated with PCOS and metabolic syndrome. The most significant changes in miRNA levels were caused by metformin treatment and the changes were seen in miRNAs previously associated with insulin resistance, obesity and liver damage, especially miR-122, miR-29a and miR-223. It appears likely that metformin, having a large effect on liver metabolism by suppressing hepatic glucose production (61), concomitantly decreases the level of several liver expressed circulating miRNAs and that this could indicate improved hepatic health.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This article is part of ReproUnion collaborative study, co-financed by the European Union, Interreg V ÔKS. The salary for P B U is funded by the grant from ReproUnion. A E S is funded by the Danish Diabetes Academy funded by Novo Nordisk Foundation. The funders had no influence on the design of the study, interpretation of results or decision to publish. The work of D G was supported by the Jacob Madsen’s and Olga Madsen’s Foundation, the Institute of Clinical Research, the Odense University Hospital, Kolding Hospital, the A. P. Møller’s Foundation, the Bernhard and Marie Kleins Foundation, The Novo Nordisk Foundation, and The Danish Medical Association. Oral contraceptive pills and metformin tablets were sponsored by Sandoz.
Acknowledgement
The authors would like to thank Christa Persson, Department of Natural Science and Environment, Roskilde University
References
- 1.Teede HJ, Misso ML, Costello MF, Dokras A, Laven J, Moran L, Piltonen T, Norman RJ.International PCOS Network. Recommendations from the international evidence-based guideline for the assessment and management of polycystic ovary syndrome. Clinical Endocrinology 2018. 89 251–2. ( 10.1111/cen.13795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burghen GA, Givens JR, Kitabchi AE.Correlation of hyperandrogenism with hyperinsulinism in polycystic ovarian disease. Journal of Clinical Endocrinology and Metabolism 1980. 50 113–11. ( 10.1210/jcem-50-1-113) [DOI] [PubMed] [Google Scholar]
- 3.Ollila M-ME, West S, Keinänen-Kiukaanniemi S, Jokelainen J, Auvinen J, Puukka K, Ruokonen A, Järvelin M-R, Tapanainen JS, Franks S.et al Overweight and obese but not normal weight women with PCOS are at increased risk of type 2 diabetes mellitus – a prospective, population-based cohort study. Human Reproduction 2016. 32 423–4. ( 10.1093/humrep/dex030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kakoly NS, Khomami MB, Joham AE, Cooray SD, Misso ML, Norman RJ, Harrison CL, Ranasinha S, Teede HJ, Moran LJ.Ethnicity, obesity and the prevalence of impaired glucose tolerance and type 2 diabetes in PCOS: a systematic review and meta-regression. Human Reproduction Update 2018. 24 455–4. ( 10.1093/humupd/dmy007) [DOI] [PubMed] [Google Scholar]
- 5.Dunaif A, Segal KR, Shelley DR, Green G, Dobrjansky A, Licholai T.Evidence for distinctive and intrinsic defects in insulin action in polycystic ovary syndrome. Diabetes 1992. 41 1257–12. ( 10.2337/diab.41.10.1257) [DOI] [PubMed] [Google Scholar]
- 6.Teede H, Tassone EC, Piltonen T, Malhotra J, Mol BW, Peña A, Witchel SF, Joham A, McAllister V, Romualdi D.et al Effect of the combined oral contraceptive pill and/or metformin in the management of PCOS: a systematic review with meta-analyses. Clinical Endocrinology 2019. 91 479–489. ( 10.1111/cen.14013) [DOI] [PubMed] [Google Scholar]
- 7.Costello M, Shrestha B, Eden J, Sjoblom P, Johnson N.Insulin-sensitising drugs versus the combined oral contraceptive pill for hirsutism, acne and risk of diabetes, cardiovascular disease, and endometrial cancer in polycystic ovary syndrome. Cochrane Database of Systematic Reviews 2007. 1 CD005552. ( 10.1002/14651858.CD005552.pub2) [DOI] [PubMed] [Google Scholar]
- 8.Rome S.Are extracellular microRNAs involved in type 2 diabetes and related pathologies? Clinical Biochemistry 2013. 46 937–9. ( 10.1016/j.clinbiochem.2013.02.018) [DOI] [PubMed] [Google Scholar]
- 9.Vienberg SG, Björnholm M.Chronic glucocorticoid treatment increases de novo lipogenesis in visceral adipose tissue. Acta Physiologica 2014. 211 257–25. ( 10.1111/apha.12283) [DOI] [PubMed] [Google Scholar]
- 10.Ono K, Igata M, Kondo T, Kitano S, Takaki Y, Hanatani S, Sakaguchi M, Goto R, Senokuchi T, Kawashima J.et al Identification of microRNA that represses IRS-1 expression in liver. PLoS ONE 2018. 13 e0191553. ( 10.1371/journal.pone.0191553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunez Lopez YO, Garufi G, Pasarica M, Seyhan AA.Elevated and correlated expressions of miR-24, miR-30d, miR-146a, and SFRP-4 in human abdominal adipose tissue play a role in adiposity and insulin resistance. International Journal of Endocrinology 2018. 2018 7351902 ( 10.1155/2018/7351902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arner P, Kulyté A.MicroRNA regulatory networks in human adipose tissue and obesity. Nature Reviews: Endocrinology 2015. 11 276–2. ( 10.1038/nrendo.2015.25) [DOI] [PubMed] [Google Scholar]
- 13.Esteves JV, Yonamine CY, Pinto-Junior DC, Gerlinger-Romero F, Enguita FJ, MacHado UF.Diabetes modulates microRNAs 29b-3p, 29c-3p, 199a-5p and 532–3p expression in muscle: possible role in GLUT4 and HK2 repression. Frontiers in Endocrinology 2018. 9 536. ( 10.3389/fendo.2018.00536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massart J, Sjögren RJO, Lundell LS, Mudry JM, Franck N, O’Gorman DJ, Egan B, Zierath JR, Krook A.Altered miR-29 expression in type 2 diabetes influences glucose and lipid metabolism in skeletal muscle. Diabetes 2017. 66 1807–18. ( 10.2337/db17-0141) [DOI] [PubMed] [Google Scholar]
- 15.Vienberg S, Geiger J, Madsen S, Dalgaard LT.MicroRNAs in metabolism. Acta Physiologica 2017. 219 346–3. ( 10.1111/apha.12681) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willeit P, Skroblin P, Kiechl S, Fernández-Hernando C, Mayr M.Liver microRNAs: potential mediators and biomarkers for metabolic and cardiovascular disease? European Heart Journal 2016. 37 3260–326. ( 10.1093/eurheartj/ehw146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guay C, Regazzi R.Circulating microRNAs as novel biomarkers for diabetes mellitus. Nature Reviews: Endocrinology 2013. 9 513–521. ( 10.1038/nrendo.2013.86) [DOI] [PubMed] [Google Scholar]
- 18.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A.et al Circulating microRNAs as stable blood-based markers for cancer detection. PNAS 2008. 105 10513–1051. ( 10.1073/pnas.0804549105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sørensen AE, Wissing ML, Salö S, Englund ALM, Dalgaard LT.MicroRNAs related to polycystic ovary syndrome (PCOS). Genes 2014. 5 684–708. ( 10.3390/genes5030684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arancio W, Calogero Amato M, Magliozzo M, Pizzolanti G, Vesco R, Giordano C.Serum miRNAs in women affected by hyperandrogenic polycystic ovary syndrome: the potential role of miR-155 as a biomarker for monitoring the estroprogestinic treatment. Gynecological Endocrinology 2018. 34 704–70. ( 10.1080/09513590.2018.1428299) [DOI] [PubMed] [Google Scholar]
- 21.Murri M, Insenser M, Fernández-Durán E, San-Millán JL, Luque-Ramírez M, Escobar-Morreale HF.Non-targeted profiling of circulating microRNAs in women with polycystic ovary syndrome (PCOS): effects of obesity and sex hormones. Metabolism: Clinical and Experimental 2018. 86 49–60. ( 10.1016/j.metabol.2018.01.011) [DOI] [PubMed] [Google Scholar]
- 22.Song DK, Sung YA, Lee H.The role of serum microRNA-6767-5p as a biomarker for the diagnosis of polycystic ovary syndrome. PLoS ONE 2016. 11 e0163756. ( 10.1371/journal.pone.0163756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willeit P, Skroblin P, Moschen AR, Yin X, Kaudewitz D, Zampetaki A, Barwari T, Whitehead M, Ramírez CM, Goedeke L.et al Circulating microRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes 2017. 66 347–3. ( 10.2337/db16-0731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghai V, Baxter D, Wu X, Kim TK, Kuusisto J, Laakso M, Connolly T, Li Y, Andrade-Gordon P, Wang K.Circulating RNAs as predictive markers for the progression of type 2 diabetes. Journal of Cellular and Molecular Medicine 2019. 23 2753–27. ( 10.1111/jcmm.14182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaeger A, Zollinger L, Saely CH, Muendlein A, Evangelakos I, Nasias D, Charizopoulou N, Schofield JD, Othman A, Soran H.et al Circulating microRNAs -192 and -194 are associated with the presence and incidence of diabetes mellitus. Scientific Reports 2018 8 1–14. ( 10.1038/s41598-018-32274-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortega FJ, Mercader JM, Moreno-Navarrete JM, Rovira O, Guerra E, Esteve E, Xifra G, Martínez C, Ricart W, Rieusset J.et al Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care 2014. 37 1375–13. ( 10.2337/dc13-1847) [DOI] [PubMed] [Google Scholar]
- 27.Ghai V, Kim TK, Etheridge A, Nielsen T, Hansen T, Pedersen O, Galas D, Wang K.Extracellular vesicle encapsulated microRNAs in patients with type 2 diabetes are affected by metformin treatment. Journal of Clinical Medicine 2019. 8 617. ( 10.3390/jcm8050617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Catanzaro G, Besharat ZM, Chiacchiarini M, Abballe L, Sabato C, Vacca A, Borgiani P, Dotta F, Tesauro M, Po A.et al Circulating microRNAs in elderly type 2 diabetic patients. International Journal of Endocrinology 2018. 2018 6872635 ( 10.1155/2018/6872635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mensà E, Giuliani A, Matacchione G, Gurău F, Bonfigli AR, Romagnoli F, De Luca M, Sabbatinelli J, Olivieri F.Circulating miR-146a in healthy aging and type 2 diabetes: age- and gender-specific trajectories. Mechanisms of Ageing and Development 2019. 180 1–10. ( 10.1016/j.mad.2019.03.001) [DOI] [PubMed] [Google Scholar]
- 30.Glintborg D, Altinok ML, Mumm H, Hermann AP, Ravn P, Andersen M.Body composition is improved during 12 months’ treatment with metformin alone or combined with oral contraceptives compared with treatment with oral contraceptives in polycystic ovary syndrome. Journal of Clinical Endocrinology and Metabolism 2014. 99 2584–25. ( 10.1210/jc.2014-1135) [DOI] [PubMed] [Google Scholar]
- 31.Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertility and Sterility 2004. 81 19–25. ( 10.1016/j.fertnstert.2003.10.004) [DOI] [PubMed] [Google Scholar]
- 32.Pfaffl MW.A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 2001. 29 e45. ( 10.1093/nar/29.9.e45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T.Identification of tissue-specific microRNAs from mouse. Current Biology 2002. 12 735–73. ( 10.1016/s0960-9822(0200809-6) [DOI] [PubMed] [Google Scholar]
- 34.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R.et al miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biology 2004. 1 106–1. ( 10.4161/rna.1.2.1066) [DOI] [PubMed] [Google Scholar]
- 35.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M.Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005. 438 685–68. ( 10.1038/nature04303) [DOI] [PubMed] [Google Scholar]
- 36.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R.et al miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metabolism 2006. 3 87–98. ( 10.1016/j.cmet.2006.01.005) [DOI] [PubMed] [Google Scholar]
- 37.Wang R, Hong J, Cao Y, Shi J, Gu W, Ning G, Zhang Y, Wang W.Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. European Journal of Endocrinology 2015. 172 291–300. ( 10.1530/EJE-14-0867) [DOI] [PubMed] [Google Scholar]
- 38.Zhu Z, Yin J, Li DCC, Mao ZQQ.Role of microRNAs in the treatment of type 2 diabetes mellitus with Roux-en-Y gastric bypass. Brazilian Journal of Medical and Biological Research 2017. 50 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunez Lopez YO, Coen PM, Goodpaster BH, Seyhan AA.Gastric bypass surgery with exercise alters plasma microRNAs that predict improvements in cardiometabolic risk. International Journal of Obesity 2017. 41 1121–11. ( 10.1038/ijo.2017.84) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hess AL, Larsen LH, Udesen PB, Sanz Y, Larsen TM, Dalgaard LT.Levels of circulating miR-122 are associated with weight loss and metabolic syndrome. Obesity 2020. 8 493–501. 10.1002/oby.22704) [DOI] [PubMed] [Google Scholar]
- 41.Noren Hooten N, Martin-Montalvo A, Dluzen DF, Zhang Y, Bernier M, Zonderman AB, Becker KG, Gorospe M, de Cabo R, Evans MK.Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence. Aging Cell 2016. 15 572–5. ( 10.1111/acel.12469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y, Gu P, Shi W, Li J, Hao Q, Cao X, Lu Q, Zeng Y.MicroRNA-29a induces insulin resistance by targeting PPARδ in skeletal muscle cells. International Journal of Molecular Medicine 2016. 37 931–93. ( 10.3892/ijmm.2016.2499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He A, Zhu L, Gupta N, Chang Y, Fang F.Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Molecular Endocrinology 2007. 21 2785–27. ( 10.1210/me.2007-0167) [DOI] [PubMed] [Google Scholar]
- 44.Pandey AK, Verma G, Vig S, Srivastava S, Srivastava AK, Datta M.miR-29a levels are elevated in the db/db mice liver and its overexpression leads to attenuation of insulin action on PEPCK gene expression in HepG2 cells. Molecular and Cellular Endocrinology 2011. 332 125–1. ( 10.1016/j.mce.2010.10.004) [DOI] [PubMed] [Google Scholar]
- 45.Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao Y, Dong Q, Pang Z, Guan Q, Gao L.et al Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetologica 2011. 48 61–6. ( 10.1007/s00592-010-0226-0) [DOI] [PubMed] [Google Scholar]
- 46.Bagge A, Clausen TR, Larsen S, Ladefoged M, Rosenstierne MW, Larsen L, Vang O, Nielsen JH, Dalgaard LT.MicroRNA-29a is up-regulated in beta-cells by glucose and decreases glucose-stimulated insulin secretion. Biochemical and Biophysical Research Communications 2012. 426 266–2. ( 10.1016/j.bbrc.2012.08.082) [DOI] [PubMed] [Google Scholar]
- 47.Dooley J, Garcia-Perez JE, Sreenivasan J, Schlenner SM, Vangoitsenhoven R, Papadopoulou AS, Tian L, Schonefeldt S, Serneels L, Deroose C.et al The microRNA-29 family dictates the balance between homeostatic and pathological glucose handling in diabetes and obesity. Diabetes 2016. 65 53–61. ( 10.2337/db15-0770) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurtz CL, Peck BCE, Fannin EE, Beysen C, Miao J, Landstreet SR, Ding S, Turaga V, Lund PK, Turner S.et al MicroRNA-29 fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes. Diabetes 2014. 63 3141–314. ( 10.2337/db13-1015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sørensen AE, Wissing ML, Englund ALM, Dalgaard LT.MicroRNA species in follicular fluid associating with polycystic ovary syndrome and related intermediary phenotypes. Journal of Clinical Endocrinology and Metabolism 2016. 101 1579–1589. ( 10.1210/jc.2015-3588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delic D, Eisele C, Schmid R, Luippold G, Mayoux E, Grempler R.Characterization of micro-RNA changes during the progression of type 2 diabetes in Zucker diabetic fatty rats. International Journal of Molecular Sciences 2016. 17 665. ( 10.3390/ijms17050665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morishita A, Yoneyama H, Iwama H, Fujita K, Watanabe M, Hirose K, Tadokoro T, Oura K, Sakamoto T, Mimura S.et al Circulating microRNA-636 is associated with the elimination of hepatitis C virus by ombitasvir/paritaprevir/ritonavir. Oncotarget 2018. 9 32054–320. ( 10.18632/oncotarget.25889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salem AM, Ragheb AS, Hegazy MGA, Matboli M, Eissa S.Caffeic acid modulates miR-636 expression in diabetic nephropathy rats. Indian Journal of Clinical Biochemistry 2018. 34 1–8. ( 10.1007/s12291-018-0743-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eissa S, Matboli M, Aboushahba R, Bekhet MM, Soliman Y.Urinary exosomal microRNA panel unravels novel biomarkers for diagnosis of type 2 diabetic kidney disease. Journal of Diabetes and its Complications 2016. 30 1585–15. ( 10.1016/j.jdiacomp.2016.07.012) [DOI] [PubMed] [Google Scholar]
- 54.Vickers KC, Landstreet SR, Levin MG, Shoucri BM, Toth CL, Taylor RC, Palmisano BT, Tabet F, Cui HL, Rye KA.et al MicroRNA-223 coordinates cholesterol homeostasis. PNAS 2014. 111 14518–145. ( 10.1073/pnas.1215767111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chuang TY, Wu HL, Chen CC, Gamboa GM, Layman LC, Diamond MP, Azziz R, Chen YH.MicroRNA-223 expression is upregulated in insulin resistant human adipose tissue. Journal of Diabetes Research 2015. 2015 943659 ( 10.1155/2015/943659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nunez Lopez YO, Garufi G, Seyhan AA.Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Molecular Biosystems 2016. 13 106–1. ( 10.1039/c6mb00596a) [DOI] [PubMed] [Google Scholar]
- 57.Wen D, Qiao P, Wang L.Circulating microRNA-223 as a potential biomarker for obesity. Obesity Research and Clinical Practice 2015. 9 398–404. ( 10.1016/j.orcp.2015.01.006) [DOI] [PubMed] [Google Scholar]
- 58.Shepherd PR, Kahn BB.Glucose transporters and insulin action – implications for insulin resistance and diabetes mellitus. New England Journal of Medicine 1999. 341 248–2. ( 10.1056/NEJM199907223410406) [DOI] [PubMed] [Google Scholar]
- 59.de Rie D, Abugessaisa I, Alam T, Arner E, Arner P, Ashoor H, Åström G, Babina M, Bertin N, Burroughs AM.et al An integrated expression atlas of miRNAs and their promoters in human and mouse. Nature Biotechnology 2017. 35 872–8. ( 10.1038/nbt.3947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bridgeman SC, Ellison GC, Melton PE, Newsholme P, Mamotte CDS.Epigenetic effects of metformin: from molecular mechanisms to clinical implications. Diabetes, Obesity and Metabolism 2018. 20 1553–15. ( 10.1111/dom.13262) [DOI] [PubMed] [Google Scholar]
- 61.An H, He L.Current understanding of metformin effect on the control of hyperglycemia in diabetes. Journal of Endocrinology 2016. 228 R97–R. ( 10.1530/JOE-15-0447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sørensen AE, Udesen PB, Maciag G, Geiger J, Saliani N, Januszewski AS, Jiang G, Ma RC, Hardikar AA.et al Hyperandrogenism and metabolic syndrome are associated with changes in serum-derived microRNAs in women with polycystic ovary syndrome. Frontiers in Medicine 6 242–6. ( 10.3389/fmed.2019.00242) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a