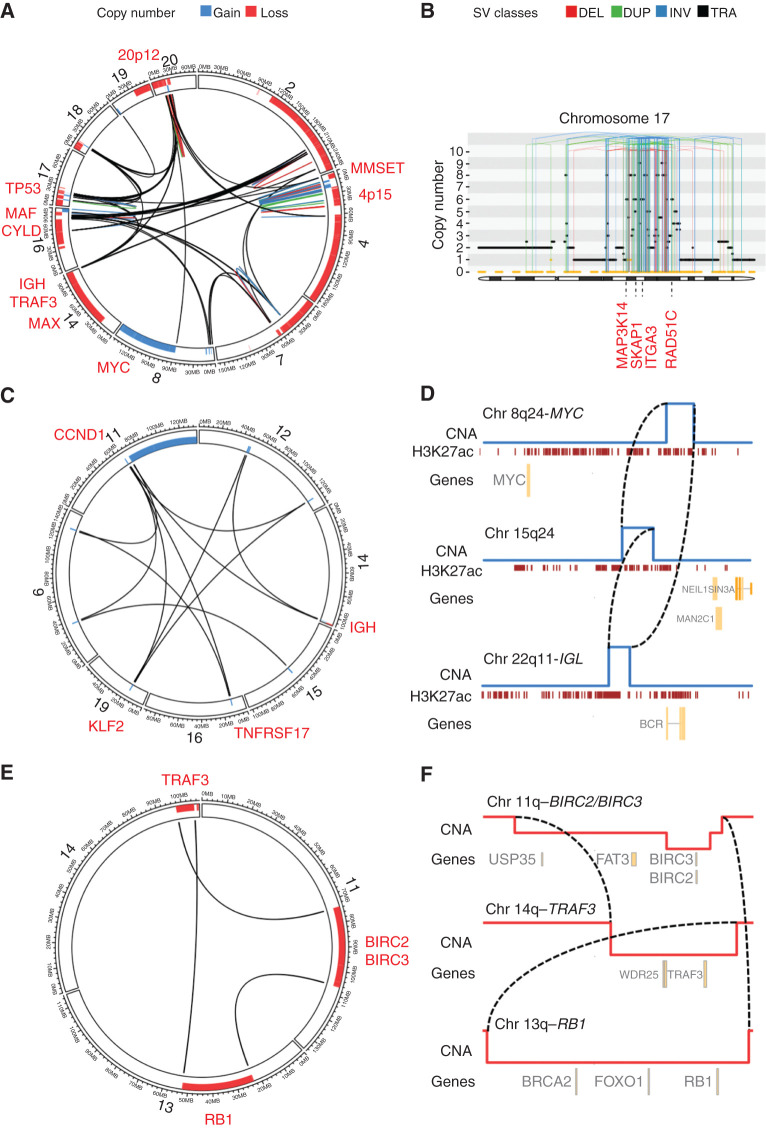
Figure 1.
Complex SV classes in multiple myeloma. A, Chromothripsis involving IGH and nine recurrent drivers across 10 different chromosomes (sample MMRF_1890_1_BM). B, Chromothripsis causing high-level focal gains on chromosome 17 (sample MMRF_2330_1_BM). The horizontal black line indicates total copy number; the dashed orange line indicates minor copy number. Vertical lines represent SV breakpoints, color-coded by SV class. Selected overexpressed genes (Z-score >2) are annotated in red, including the established multiple myeloma driver gene MAP3K14 and RAD51C, an oncogene commonly amplified in breast cancer (ref. 66; six copies). C, Templated insertion involving seven different chromosomes, causing a canonical IGH-CCND1 translocation and involving at least two additional drivers in the same event (i.e., KLF2 and TNFRSF17; sample MMRF_1677_1_BM). D, Simpler templated insertion cycle (brown lines), involving IGL, MYC, and a hotspot on chromosome 15q24 (sample MMRF_1550_1_BM). Copy-number profile shown in blue, with active enhancers below in brown (H3K27Ac). E, Chromoplexy involving chromosomes 11, 13, and 14, simultaneously causing deletion of key tumor suppressor genes on each chromosome (sample MMRF_2194_1_BM). F, Zooming in on the translocations and associated large deletions, which make up the chromoplexy event depicted as a Circos plot in C; (sample MMRF_2194_1_BM). The Circos plots in panels A, C, and E each show the SV breakpoints of a single complex SV (colored lines; legend above panels), with bars around the plot circumference indicating copy-number changes (red, loss; blue, gain).