Figure 2. scATAC-Seq Reveals Open Chromatin Landscapes of Single Cells in Neonatal Hearts.
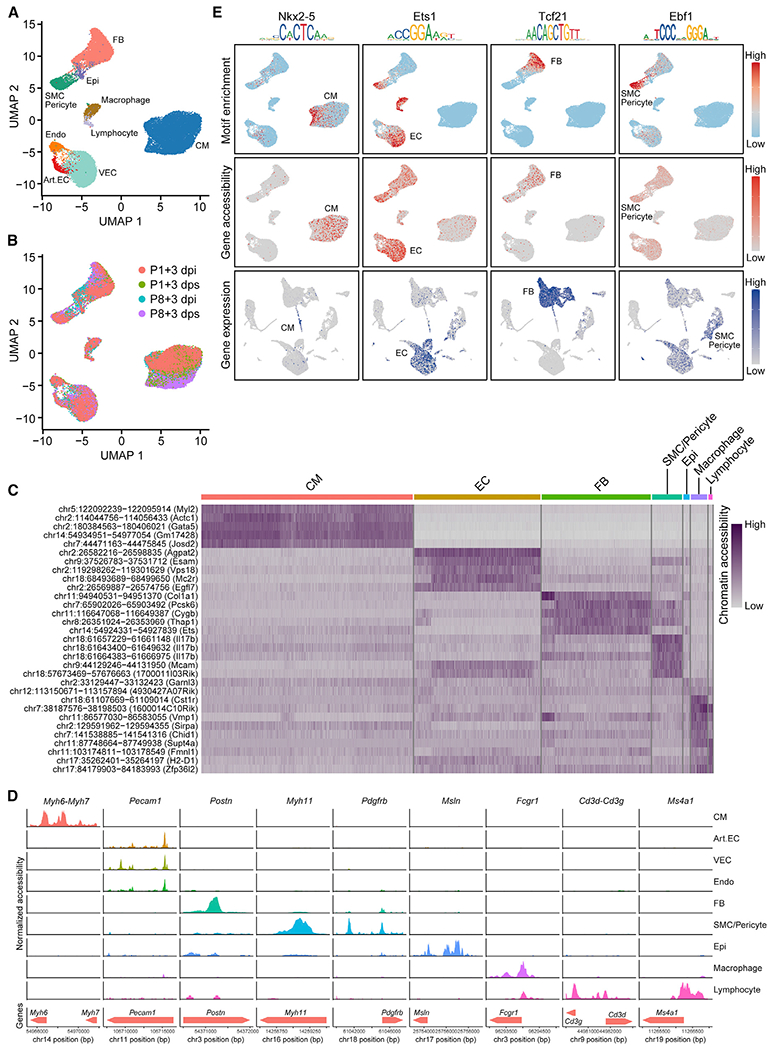
(A and B) UMAP plots showing single-cell open chromatin profiles analyzed in the study, color coded for cell clusters (A) or sample origins (B). For each time point, one scATAC-seq library was generated using pooled tissues dissected from 8–12 individual animals to control the differences among individual animals and dissection variations.
(C) Heatmap showing activity of top enriched open chromatin peaks and their associated genomic coordinates for each cell cluster.
(D) scATAC-seq tracks showing open chromatin peaks associated with cell-type-specific genes, including Myh6 and Myh7 (CM markers), Pecam1 (EC marker), Postn (FB marker), Myh11 (SMC marker), Pdgfrb (pericyte marker), Msln (epicardial cell marker), Fcgr1 (macrophage marker), Cd3g and Cd3d (T cell markers), and Ms4a1 (B cell marker), across different cell clusters.
(E) Transcription factor motif enrichment (upper row), gene accessibility (middle row), and gene expression profiles (bottom row) for lineage-specific transcription factors Nkx2–5 (CM specific, first column), Ets1 (EC enriched, second column), Tcf21 (FB specific, third column), and Ebf1 (SMC/Pericyte enriched, fourth column).
Art.EC, arterial endothelial cell; CM, cardiomyocyte; dpi, days postinjury; dps, days post sham; Endo, endocardial cell; Epi, epicardial cell; FB, fibroblast; SMC, smooth muscle cell; VEC, vascular endothelial cell. See also Figures S2 and S3 and Tables S1 and S2.
