Fig 3. Characterization of K25N HIV-1 and K25N/E45A HIV-1.
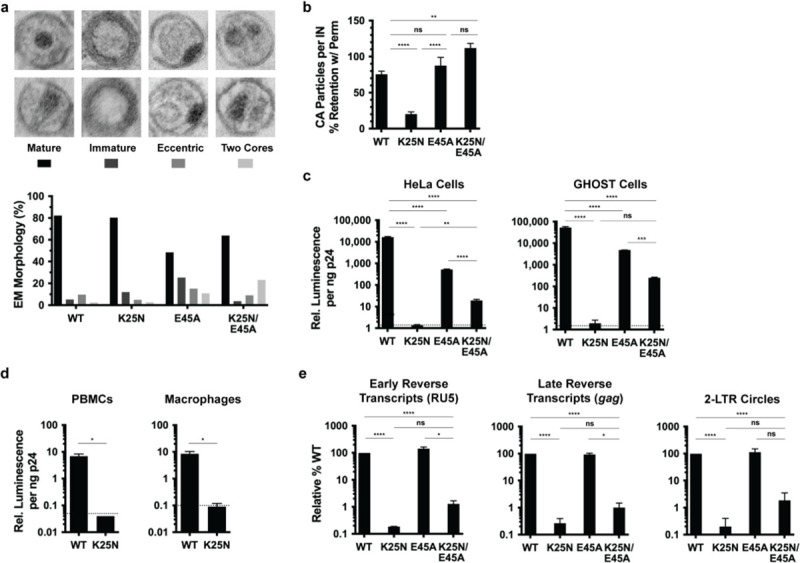
(a) TEM classification of virus particle morphologies for HIV-1NL4-3 bearing indicated CA changes are shown for 2 independent experiments, with >200 particles counted per experiment for each virus type (>480 particles of WT and each mutant virus across experiments). Representative images of the different morphologies are shown for illustrative purposes. Statistical significance between WT and mutant morphologies are indicated. (b) Viruses bearing the indicated CA substitutions were captured on glass with or without virus membrane permeabilization, followed by immunostaining for CA and imaging. Data are expressed as the percentage of non-permeabilized CA staining retained with membrane permeabilization for each virus. (c, d) Single-cycle infectivities of WT and indicated CA mutant viruses in indicated cells. Dotted lines represent average luciferase signal of uninfected cells. (e) DNA synthesis and 2-LTR circle formation of WT and CA mutant viruses. Error bars indicate SEM for 2–4 experiments. **** P < 0.0001, *** P < 0.001, ** P < 0.01, * P < 0.05. Numerical data for panels A, B, C, D, and E can be found in S3 Data. 2-LTR, 2-long terminal repeat; TEM, transmission electron microscopy; IN, integrase; ns, not significant; PBMC, peripheral blood mononuclear cell; WT, wild-type.
